.svg) National Institute of General Medical Sciences |
 |
 |
National Biomedical Resource for |
| ACERT's Service and Collaborative Projects | |
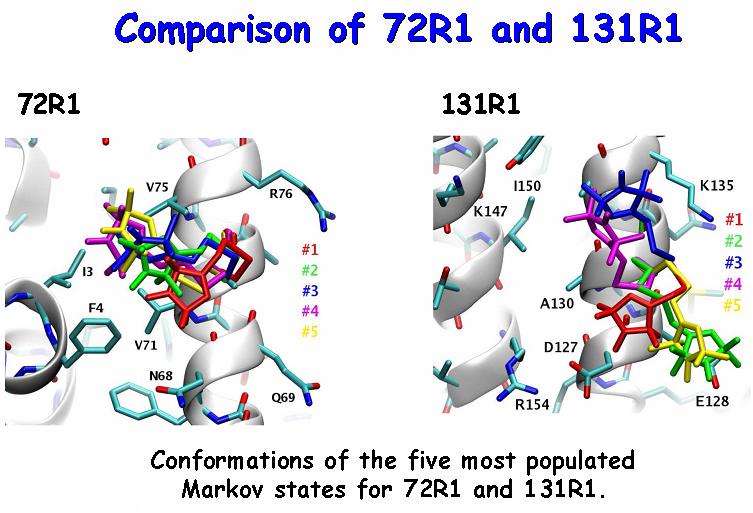
ESR in combination with site-directed spin labeling is a powerful biophysical technique for probing protein conformation and its changes during function in native, aqueous or membrane environments. When a nitroxide spin label is attached to all the possible positions along the protein sequence, overall trends and general patterns across the recorded spectra provide information about the secondary structure of the protein and, when used with other paramagnetic species, about the orientation of its subunits relative to each other or to the solvent. The specific features of each spectral line shape need to be carefully understood and interpreted when detailed information is sought. Since the local protein dynamics and structure are manifested in the ESR spectrum through their modulation of the internal spin label dynamics, close familiarity with the behavior of the reporter itself becomes necessary. Multifrequency electron spin resonance (ESR) spectra contain very rich information about the structure and dynamics of the local environment of the spin label. Relating the features of the observed spectra to the molecular motion and interactions that cause them, however, has proven to be challenging. We have performed extensive molecular dynamics simulations of fully solvated T4 Lysozyme, spin labeled at positions 72 and 131. The atomistic trajectories were then utilized to construct Markov chain models of the internal spin label dynamics. Combined with rotational diffusion of the global protein tumbling, the Markov jump trajectories were used to simulate multifrequency ESR spectra, which are in quantitative agreement with experiment. The conformations of the most probable Markov states (cf. Figure) and the energies of their interaction with the protein surface were also obtained. Our results demonstrate that the behavior at positions 72 and 131 is more complex than commonly believed. Due to its amphipathic nature, the nitroxide ring of the spin label appears to utilize every possibility to establish van der Waals contacts with a hydrophobic residue or form a salt bridge with a charged one. The length of the linker allows R1 to extend its range of exploration beyond its immediate neighbors. Given the differences between 72R1 and 131R1 (cf. Figure) it is likely that the coupling between the nitroxide linker and the protein backbone fluctuations helps to drive the conformational dynamics of the linker. The match between the calculated and the experimental spectra at three different frequencies (and corresponding magnetic fields) provides strong support for the relevance of the MD simulations and their subsequent analysis. Publication: D. Sezer, J.H. Freed, and B. Roux, J. Am. Chem. Soc., 131, 2597-2605 (2009); PMC2763593 |
|
 |
|
|
D. Sezer, (Department of Physics and Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, Illinois) J. H. Freed, (ACERT) B. Roux (Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, Illinois) |
|
|
|
About ACERT Contact Us |
Research |
Outreach |
ACERT is supported by grant 1R24GM146107 from the National Institute of General Medical Sciences (NIGMS), part of the National Institutes of Health. |
|||||
| ||||||||