.svg) National Institute of General Medical Sciences |
 |
 |
National Biomedical Resource for |
| ACERT's Service and Collaborative Projects | |
Protein structure determination by spectroscopic methods requires measurement of distances relevant to the macromolecular structure. These distances serve as constraints in computational procedures to reduce the number of degrees of freedom available to the polypeptide chain, to orient and dock proteins that participate in supramolecular assemblies, and to characterize the time evolution of these structures during their function. Efforts to develop approaches for distance measurements are particularly timely in the context of structural genomics for problems that are currently not amenable to study by X-ray crystallography. Notable examples include the static structure of membrane proteins and conformationally heterogeneous water soluble proteins and the fluctuations in these structures. In this context, ESR spectroscopy in combination with site-directed spin labeling provides unique capabilities. We have used a novel pulsed ESR technique for distance measurement, based on the detection of double quantum coherence (DQC), which yields high quality dipolar spectra, to significantly extend the range of measurable distances in proteins using nitroxide spin-labels beyond what is possible with conventional ESR. T4 lysozyme mutants, double labeled with MTSSL spin label have been studied using DQC-ESR. The distances span the range from 20 to 47 Å. The high quality of the dipolar spectra also yields distance distributions, which can be used to set upper and lower bounds. The shape of these distributions can reveal the presence of multiple conformations of the spin label, important for the interpretation of the distances. The distances and distributions we found are readily rationalized in terms of the known crystal structure, the characteristic conformers of the nitroxide side-chains, and molecular modeling. We show in the figure below how our approach enables "triangulating" the tertiary structure. The triangulated distances shown are from the modeling and are in good agreement with the experimental results. This study sets the stage for the use of DQC-ESR for determining the tertiary structure of large proteins with just a small number of long-distance constraints. Publication: P.P. Borbat, H.S. Mchaourab, and J.H. Freed, J. Am. Chem. Soc., 124, 5304-5314 (2002); no PMCID |
|
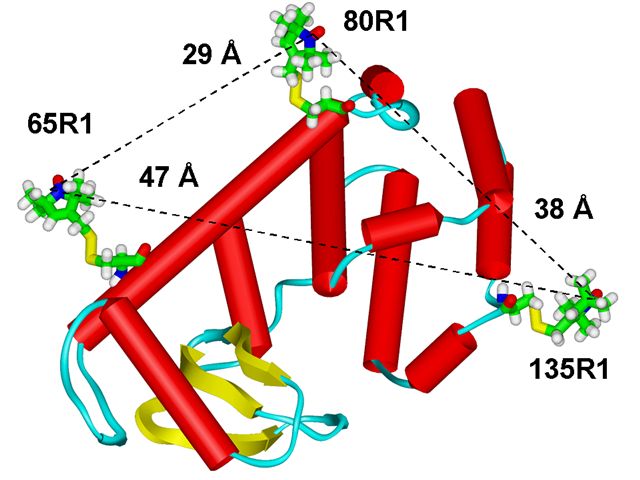 |
|
|
P. Borbat, J. Freed (ACERT), H. Mchaourab (Vanderbilt University) |
|
|
|
About ACERT Contact Us |
Research |
Outreach |
ACERT is supported by grant 1R24GM146107 from the National Institute of General Medical Sciences (NIGMS), part of the National Institutes of Health. |
|||||
| ||||||||