|
Flavoproteins as native and genetically encoded spin probes for in cell ESR spectroscopy
T. Chauviré, S. Chandrasekaran, R. Dunleavy, J. H. Freed, B. R. Crane
Nat. Commun. 16, 5406 (2025).
Supporting Information
<doi: 10.1038/s41467-025-60623-6>
PMID:
40595551
PMCID:
PMC12214676
Publication #469
|
|
|
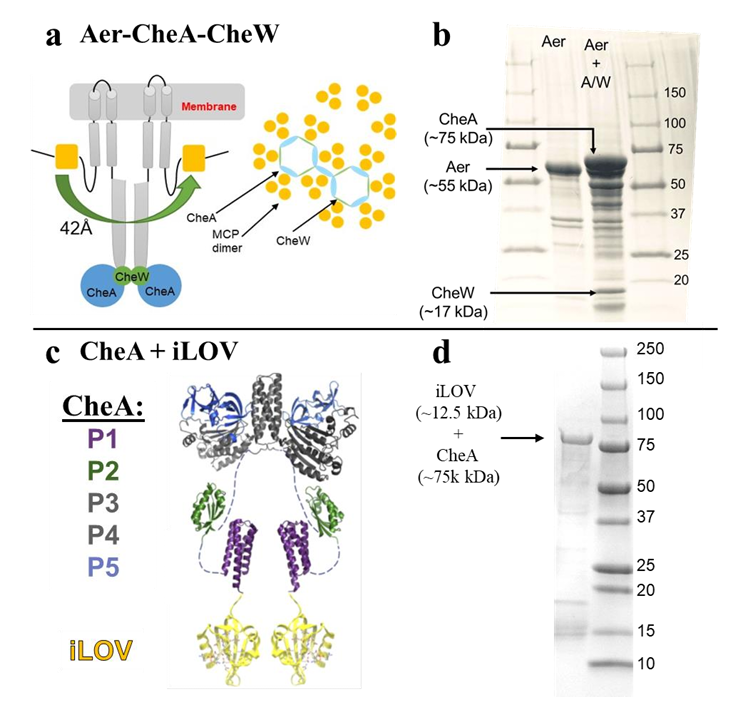
ABSTRACT: Flavin cofactors are attractive Electron Spin Resonance (ESR) probes for proteins because cellular reductants and light can generate their semiquinone states. We have used ESR spectroscopy to study the bacterial transmembrane aerotaxis receptor (Aer) in its native Escherichia coli membrane environment. Optimization of the spectroscopic (electronic relaxation times) and cell growth (isotopic labeling) conditions allowed for measurements of Aer with its partners - the histidine kinase (CheA) and the coupling protein (CheW) - in native signaling arrays. Continuous-wave ESR measurements at room temperature showed a rigid Aer flavin immobilized in the cofactor pocket and Q-band electron nuclear double resonance (ENDOR) measurements identified a predominant anionic semiquinone radical state in cell. Q-band four-pulse double electron-electron resonance (4P-DEER) measurements indicated a 4.1 nm distance between the two flavins of an Aer homodimer, consistent with previous in vitro measurements, but also revealed additional separations in cell indicative of chemoreceptor arrays, not previously observed for Aer. For general application, we further developed a genetically encoded Light-Oxygen and Voltage (LOV) domain for incorporation into target proteins as an ESR probe of structural properties in cell. This approach provides a framework to elucidate protein oligomeric states and conformations that are difficult to reproduce in vitro.
|
|
|
Rapid Analysis of DEER Signals Including Short Distances
A. Sinha Roy, T. E. Assafa, B. Dzikovski, N. Joshi, J. H. Freed
J. Phys. Chem. Lett. 16, (1) 38-44 (2025)
|
|
|
Rapid Analysis of DEER Signals Including Short Distances
A. Sinha Roy, T. E. Assafa, B. Dzikovski, N. Joshi, J. H. Freed
J. Phys. Chem. Lett. 16, (1) 38-44 (2025)
<doi: 10.1021/acs.jpclett.4c03245>
PMID:
39693563
PMCID:
PMC11717586
Publication #468
|

|
|
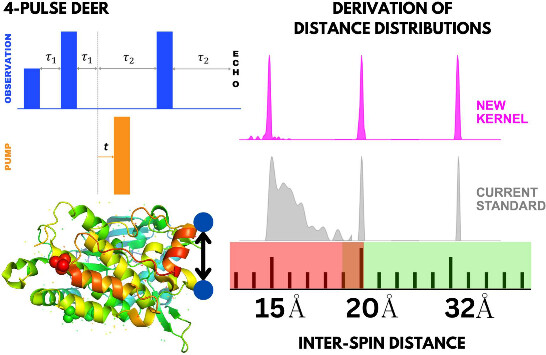
ABSTRACT: Double electron electron resonance (DEER) spectroscopy is an important technique to measure distance distributions P(r) for studying protein structures and protein–protein interactions. DEER data analysis can at times become challenging due to the lack of a detailed analytical signal expression or numerical methods with rapid computation time. We have derived an analytical expression κFULL, which includes both the pseudo-secular dipolar coupling (PSDC) and the finite pulse effects, especially important for shorter distances. Analyses of experiments by κFULL yield accurate and consistent P(r) values for three DEER nitroxide-rulers with distances (rAVG) in the range of 15 to 32 Å, while the current standard analysis produces erroneous results for rAVG < 20 Å. Computation times for deriving P(r) vary between 1 min and 4 min, which is usually much shorter than previous methods that include pseudo-secular and other effects. The expression can be applied to all types of DEER spin probes with little or no modifications.
|
|
|
Novel requirements for HAP2/GCS1-mediated gamete fusion in Tetrahymena
J. F. Pinello, J. Loidl, E. S. Seltzer, D. Cassidy-Hanley, D. Kolbin, A. Abdelatif, F. A. Rey, R. An, N. J. Newberger, Y. Bisharyan, H. Papoyan, H. Byun, H. C. Aguilar, A. L. Lai, J. H. Freed, T. Maugel, E. S. Cole, and T. G. Clark
iScience 27, 110146 (2024)
|

|
|
Novel requirements for HAP2/GCS1-mediated gamete fusion in Tetrahymena
J. F. Pinello, J. Loidl, E. S. Seltzer, D. Cassidy-Hanley, D. Kolbin, A. Abdelatif, F. A. Rey, R. An, N. J. Newberger, Y. Bisharyan, H. Papoyan, H. Byun, H. C. Aguilar, A. L. Lai, J. H. Freed, T. Maugel, E. S. Cole, and T. G. Clark
iScience 27, 110146 (2024)
Supporting Information
<doi: 10.1016/j.isci.2024.110146>
PMID:
38904066
PMCID:
PMC11187246
Publication #467
|

|
|
ABSTRACT: The ancestral gamete fusion protein, HAP2/GCS1, plays an essential role in fertilization in a broad range of taxa. To identify factors that may regulate HAP2/GCS1 activity, we screened mutants of the ciliate Tetrahymena thermophila for behaviors that mimic Δhap2/gcs1 knockout phenotypes in this species. Using this approach, we identified two new genes, GFU1 and GFU2, whose products are necessary for membrane pore formation following mating type recognition and adherence. GFU2 is predicted to be a single-pass transmembrane protein, while GFU1, though lacking obvious transmembrane domains, has the potential to interact directly with membrane phospholipids in the cytoplasm. Like Tetrahymena HAP2/GCS1, expression of GFU1 is required in both cells of a mating pair for efficient fusion to occur. To explain these bilateral requirements, we propose a model that invokes cooperativity between the fusion machinery on apposed membranes of mating cells and accounts for successful fertilization in Tetrahymena's multiple mating type system.
|
|
|
MRI Denoising Using Pixel-Wise Threshold Selection
N. Srivastava, G.R. Sahoo, H. U. Voss, S. N. Niogi, J. H. Freed, and M. Srivastava
IEEE Access 12 135730-135745 (2024)
<doi: 10.1109/ACCESS.2024.3449811>
PMID:
39640512
PMCID:
PMC11619618
Publication #466
|
|
|
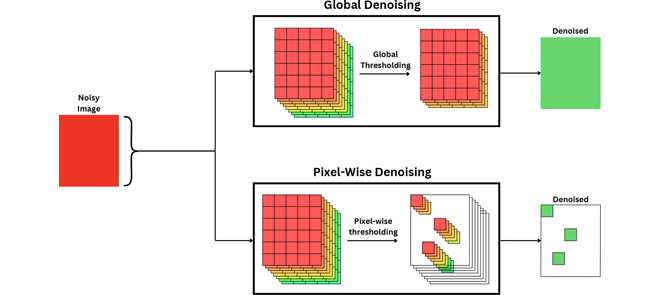
ABSTRACT: Magnetic resonance imaging (MRI) has emerged as a promising technique for non-invasive medical imaging. The primary challenge in MRI is the trade-off between image visual quality and acquisition time. Current MRI image denoising algorithms employ global thresholding to denoise the whole image, which leads to inadequate denoising or image distortion. This study introduces a novel pixel-wise (localized) thresholding approach of singular vectors, obtained from singular value decomposition, to denoise magnetic resonance (MR) images. The pixel-wise thresholding of singular vectors is performed using separate singular values as thresholds at each pixel, which is advantageous given the spatial noise variation throughout the image. The method presented is validated on MR images of a standard phantom approved by the magnetic resonance accreditation program (MRAP). The denoised images display superior visual quality and recover minute structural information otherwise suppressed in the noisy image. The increase in peak-signal-to-noise-ratio (PSNR) and contrast-to-noise-ratio (CNR) values of ≥ 18% and ≥ 200% of the denoised images, respectively, imply efficient noise removal and visual quality enhancement. The structural similarity index (SSIM) of ≥ 0.95 for denoised images indicates that the crucial structural information is recovered through the presented method. A comparison with the standard filtering methods widely used for MRI denoising establishes the superior performance of the presented method. The presented pixel-wise denoising technique reduces the scan time by 2-3 times and has the potential to be integrated into any MRI system to obtain faster and better quality images.
|
|
|
Optimal Wavelet Selection for Signal Denoising
G. R. Sahoo, J. H. Freed, M. Srivastava
IEEE Access 12 45369-45380 (2024)
<doi: 10.1109/ACCESS.2024.3377664>
PMID:
39421805
PMCID:
PMC11486496
Publication #465
|
|
|
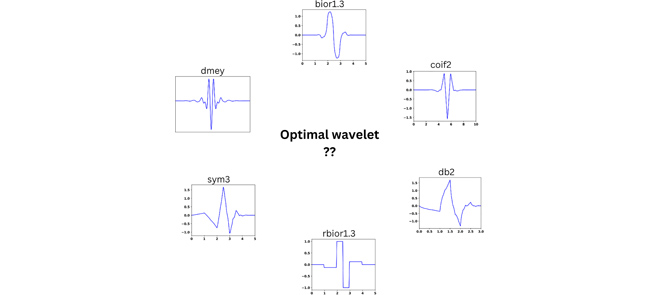
ABSTRACT: Wavelet denoising plays a key role in removing noise from signals and is widely used in many applications. In denoising, selection of the mother wavelet is desirable for maximizing the separation of noise and signal coefficients in the wavelet domain for effective noise thresholding. At present, wavelet selection is carried out in a heuristic manner or using a trial-and-error that is time consuming and prone to error, including human bias. This paper introduces a universal method to select optimal wavelets based on the sparsity of Detail components in the wavelet domain, an empirical approach. A mean of sparsity change ( μsc ) parameter is defined that captures the mean variation of noisy Detail components. The efficacy of the presented method is tested on simulated and experimental signals from Electron Spin Resonance spectroscopy at various SNRs. The results reveal that the μsc values of signal vary abruptly between wavelets, whereas for noise it displays similar values for all wavelets. For low Signal-to-Noise Ratio (SNR) data, the change in μsc between highest and second highest value is ≈8–10% and for high SNR data it is around 5%. The mean of sparsity change increases with the SNR of the signal, which implies that multiple wavelets can be used for denoising a signal, whereas, the signal with low SNR can only be efficiently denoised with a few wavelets. Either a single wavelet or a collection of optimal wavelets (i.e., top five wavelets) should be selected from the highest μsc values. The code is available on GitHub and the signalsciencelab.com website.
|
|
|
The crystal structure of bacteriophage λ RexA provides novel insights into the DNA binding properties of Rex-like phage exclusion proteins
M. C. Adams, C. J. Schiltz, J. Sun, C. J. Hosford, V. M. Johnson, H. Pan, P. P. Borbat, J. H. Freed, L. C. Thomason, C. Court, D. L. Court, J. S. Chappie
Nucleic Acids Res. 52, 4659-4675 (2024)
|

|
|
The crystal structure of bacteriophage λ RexA provides novel insights into the DNA binding properties of Rex-like phage exclusion proteins
M. C. Adams, C. J. Schiltz, J. Sun, C. J. Hosford, V. M. Johnson, H. Pan, P. P. Borbat, J. H. Freed, L. C. Thomason, C. Court, D. L. Court, J. S. Chappie
Nucleic Acids Res. 52, 4659-4675 (2024)
Supporting Information
<doi: 10.1093/nar/gkae212>
PMID:
38554102
PMCID:
PMC11077077
Publication #464
|

|
|
ABSTRACT: RexA and RexB function as an exclusion system that prevents bacteriophage T4rII mutants from growing on Escherichia coli λ phage lysogens. Recent data established that RexA is a non-specific DNA binding protein that can act independently of RexB to bias the λ bistable switch toward the lytic state, preventing conversion back to lysogeny. The molecular interactions underlying these activities are unknown, owing in part to a dearth of structural information. Here, we present the 2.05-Å crystal structure of the λ RexA dimer, which reveals a two-domain architecture with unexpected structural homology to the recombination-associated protein RdgC. Modelling suggests that our structure adopts a closed conformation and would require significant domain rearrangements to facilitate DNA binding. Mutagenesis coupled with electromobility shift assays, limited proteolysis, and double electron–electron spin resonance spectroscopy support a DNA-dependent conformational change. In vivo phenotypes of RexA mutants suggest that DNA binding is not a strict requirement for phage exclusion but may directly contribute to modulation of the bistable switch. We further demonstrate that RexA homologs from other temperate phages also dimerize and bind DNA in vitro. Collectively, these findings advance our mechanistic understanding of Rex functions and provide new evolutionary insights into different aspects of phage biology.
|
|
|
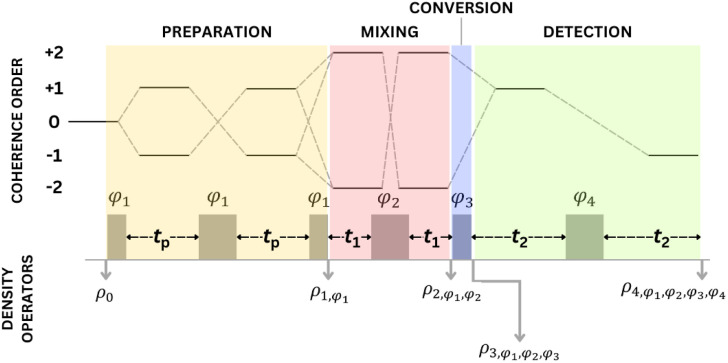
An analysis of double-quantum coherence ESR in an N-spin system: Analytical expressions and predictions
A. Sinha Roy, J. A. Marohn, J. H. Freed
J. Chem. Phys. 160, 134105 (2024)
|
|
|
An analysis of double-quantum coherence ESR in an N-spin system: Analytical expressions and predictions
A. Sinha Roy, J. A. Marohn, J. H. Freed
J. Chem. Phys. 160, 134105 (2024)
<doi: 10.1063/5.0200054>
PMID:
38557852
PMCID:
PMC11087869
Publication #463
|

|
|
ABSTRACT: Electron spin resonance pulsed dipolar spectroscopy (PDS) has become popular in protein 3D structure analysis. PDS studies yield distance distributions between a pair or multiple pairs of spin probes attached to protein molecules, which can be used directly in structural studies or as constraints in theoretical predictions. Double-quantum coherence (DQC) is a highly sensitive and accurate PDS technique to study protein structures in the solid state and under physiologically relevant conditions. In this work, we have derived analytical expressions for the DQC signal for a system with N-dipolar coupled spin-1/2 particles in the solid state. The expressions are integrated over the relevant spatial parameters to obtain closed form DQC signal expressions. These expressions contain the concentration-dependent "instantaneous diffusion" and the background signal. For micromolar and lower concentrations, these effects are negligible. An approximate analysis is provided for cases of finite pulses. The expressions obtained in this work should improve the analysis of DQC experimental data significantly, and the analytical approach could be extended easily to a wide range of magnetic resonance phenomena.
|
|
|
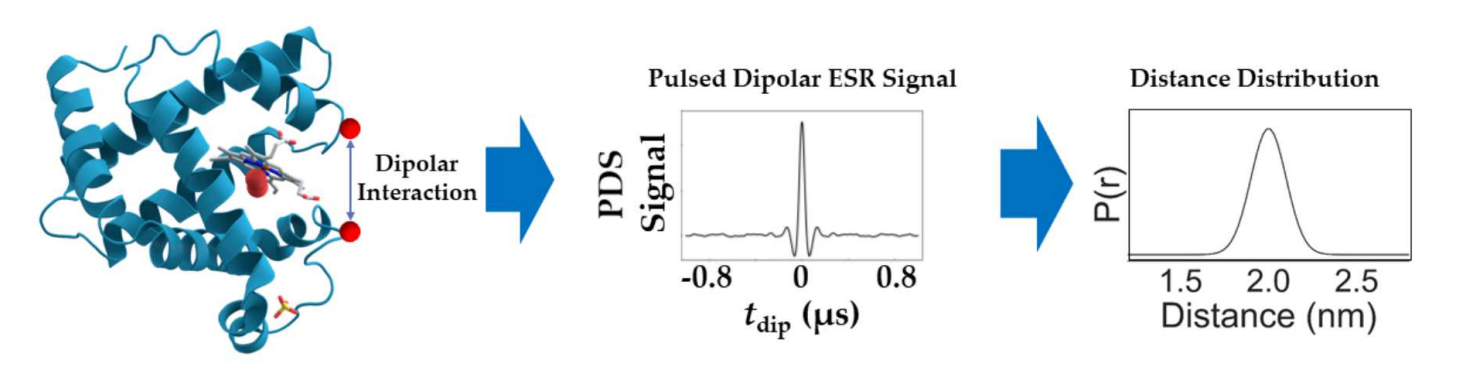
Differentiating Unimodal and Multimodal Distributions in Pulsed Dipolar Spectroscopy Using Wavelet Transforms
A.Sinha Roy, J. H. Freed, M. Srivastava
Appl. Magn. Reson. 55 (1-3) 219-237 (2024)
Supporting Information
<doi: 10.1007/s00723-023-01616-w>
PMID:
37577617
PMCID:
PMC10418556
Publication #462
|
|
|
ABSTRACT: Site directed spin labeling has enabled protein structure determination using electron spin resonance (ESR) pulsed dipolar spectroscopy (PDS). Small details in a distance distribution can be key to understanding important protein structure-function relationships. A major challenge has been to differentiate unimodal and overlapped multimodal distance distributions. They often yield similar distributions and dipolar signals. Current model-free distance reconstruction techniques such as Srivastava-Freed Singular Value Decomposition (SF-SVD) and Tikhonov regularization can suppress these small features in uncertainty and/or error bounds, despite being present. In this work, we demonstrate that continuous wavelet transform (CWT) can distinguish PDS signals from unimodal and multimodal distance distributions. We show that periodicity in CWT representation reflects unimodal distributions, which is masked for multimodal cases. This work is meant as a precursor to a cross-validation technique, which could indicate the modality of the distance distribution.
|
|
|
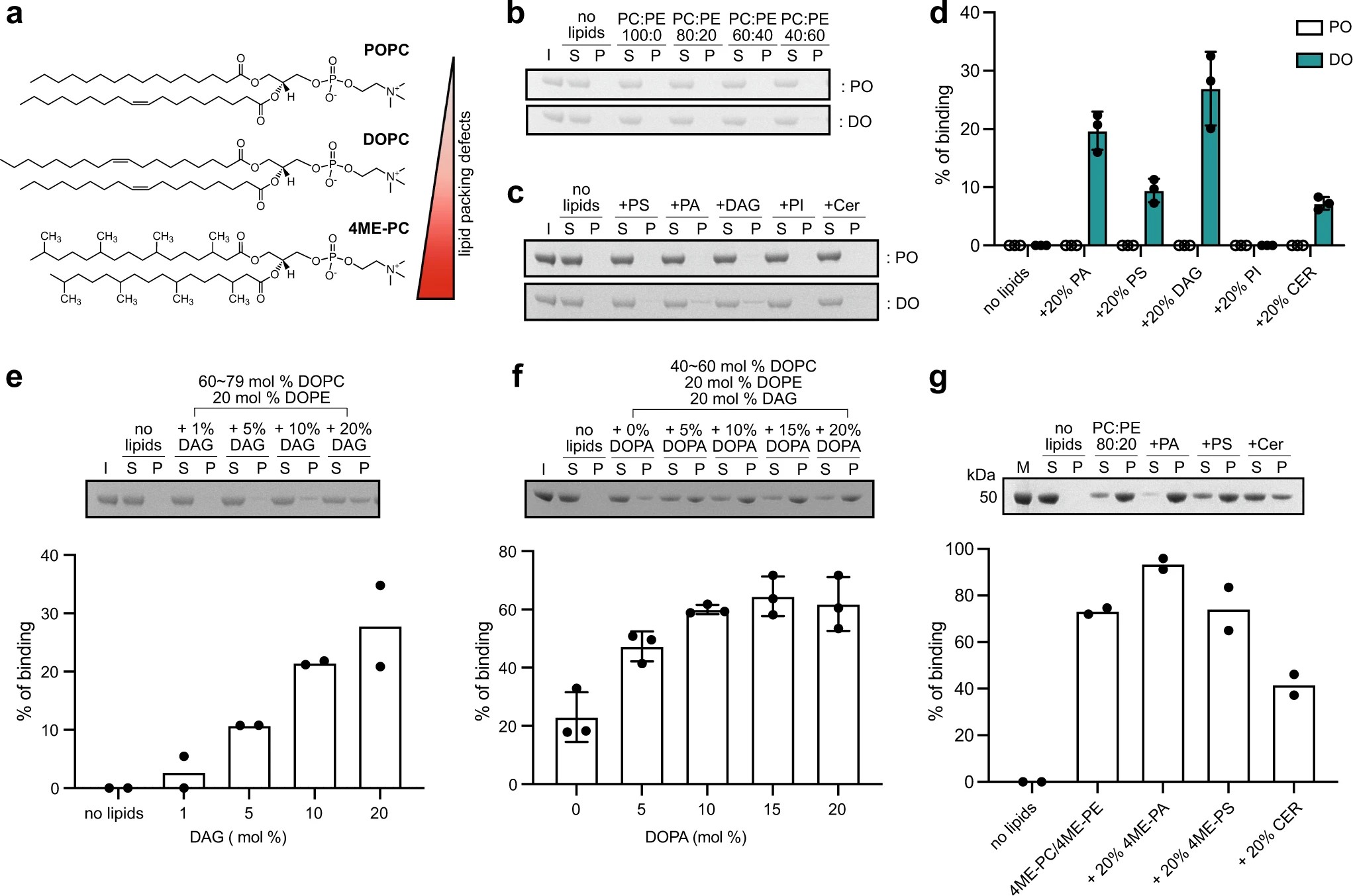
Structural insights into perilipin 3 membrane association in response to diacylglycerol accumulation
Y. M. Choi, D. Ajjaji, K. D. Fleming, P. P. Borbat, M. L. Jenkins, B. E. Moeller, S. Fernando, S. R. Bhatia, J. H. Freed, J. E. Burke, A. R. Thiam, M. V. Airola
Nat. Commun. 14 3204 (2023)
|
|
|
Structural insights into perilipin 3 membrane association in response to diacylglycerol accumulation
Y. M. Choi, D. Ajjaji, K. D. Fleming, P. P. Borbat, M. L. Jenkins, B. E. Moeller, S. Fernando, S. R. Bhatia, J. H. Freed, J. E. Burke, A. R. Thiam, M. V. Airola
Nat. Commun. 14 3204 (2023)
Supporting Information
<doi: 10.1038/s41467-023-38725-w>
PMID:
37268630
PMCID:
PMC10238389
Publication #461
|

|
|
ABSTRACT: Lipid droplets (LDs) are dynamic organelles that contain an oil core mainly composed of triglycerides (TAG) that is surrounded by a phospholipid monolayer and LD-associated proteins called perilipins (PLINs). During LD biogenesis, perilipin 3 (PLIN3) is recruited to nascent LDs as they emerge from the endoplasmic reticulum. Here, we analyze how lipid composition affects PLIN3 recruitment to membrane bilayers and LDs, and the structural changes that occur upon membrane binding. We find that the TAG precursors phosphatidic acid and diacylglycerol (DAG) recruit PLIN3 to membrane bilayers and define an expanded Perilipin-ADRP-Tip47 (PAT) domain that preferentially binds DAG-enriched membranes. Membrane binding induces a disorder to order transition of alpha helices within the PAT domain and 11-mer repeats, with intramolecular distance measurements consistent with the expanded PAT domain adopting a folded but dynamic structure upon membrane binding. In cells, PLIN3 is recruited to DAG-enriched ER membranes, and this requires both the PAT domain and 11-mer repeats. This provides molecular details of PLIN3 recruitment to nascent LDs and identifies a function of the PAT domain of PLIN3 in DAG binding.
|
|
|
Thermal degradation of thaumatin at low pH and its prevention using alkyl gallates
B. Pomon, Y. Zhao, A. L. Lai, T. Lin, J. H. Freed, A. Abbaspourrad
Food Hydrocoll. 139, 108544 (2023)
Supporting Information
<doi: 10.1016/j.foodhyd.2023.108544>
PMID:
37546699
PMCID:
PMC10399911
Publication #460
|

|
|
ABSTRACT: Thaumatin, a potent sweet tasting protein extracted from the Katemfe Plant, is emerging as a natural alternative to synthetic non-nutritive sweeteners and flavor enhancer. As a food additive, its stability within the food matrix during thermal processing is of great interest to the food industry. When heated under neutral or basic conditions, thaumatin was found to lose its sweetness due to protein aggregation caused by sulfhydryl catalyzed disulfide bond interchange. At lower pH, while thaumatin was also found to lose sweetness after heating, it does so at a slower rate and shows more resistance to sweetness loss. SDS-PAGE indicated that thaumatin fragmented into multiple smaller pieces under heating in acidic pH. Using BEMPO-3, a lipophilic spin trap, we were able to detect the presence of a free-radical within the hydrophobic region of the protein during heating. Protein carbonyl content, a byproduct of protein oxidation, also increased upon heating, providing additional evidence for protein cleavage by a radical pathway. Hexyl gallate successfully inhibited the radical generation as well as protein carbonyl formation of thaumatin during heating.
|
|
|
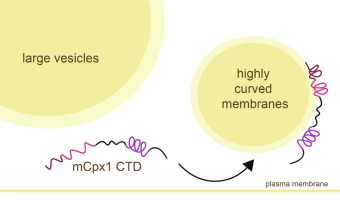
Membrane Binding Induces Distinct Structural Signatures in the Mouse Complexin–1C–Terminal Domain
E. M. Grasso, M. S. Terakawa, A. L. Lai, Y. X. Xie, T. F. Ramlall, J. H. Freed, and D. Eliezer.
J. Mol. Biol. 435, 167710 (2023)
Supporting Information
<doi: 10.1016/j.jmb.2022.167710>
PMID:
35777466
PMCID:
PMC9794636
Publication #459
|
|
|
ABSTRACT: Complexins play a critical role in regulating SNARE-mediated exocytosis of synaptic vesicles. Evolutionary divergences in complexin function have complicated our understanding of the role these proteins play in inhibiting the spontaneous fusion of vesicles. Previous structural and functional characterizations of worm and mouse complexins have indicated the membrane curvature-sensing C-terminal domain of these proteins is responsible for differences in inhibitory function. We have characterized the structure and dynamics of the mCpx1 CTD in the absence and presence of membranes and membrane mimetics using NMR, ESR, and optical spectroscopies. In the absence of lipids, the mCpx1 CTD features a short helix near its N-terminus and is otherwise disordered. In the presence of micelles and small unilamellar vesicles, the mCpx1 CTD forms a discontinuous helical structure in its C-terminal 20 amino acids, with no preference for specific lipid compositions. In contrast, the mCpx1 CTD shows distinct compositional preferences in its interactions with large unilamellar vesicles. These studies identify structural divergences in the mCpx1 CTD relative to the wCpx1 CTD in regions that are known to be critical to the wCpx1 CTD's role in inhibiting spontaneous fusion of synaptic vesicles, suggesting a potential structural basis for evolutionary divergences in complexin function.
|
|
|
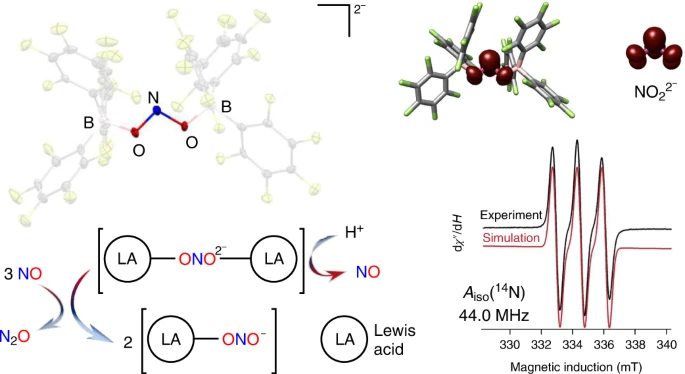
Lewis acid-assisted reduction of nitrite to nitric and nitrous oxides via the elusive nitrite radical dianion
V. Hosseininasab, I. M DiMucci, P. Ghosh, J. A. Bertke, S. Chandrasekharan, C. J. Titus, D. Nordlund, J. H. Freed, K. M. Lancaster, T. H. Warren
Nat. Chem. 14, 1265-1269 (2022)
|
|
|
Lewis acid-assisted reduction of nitrite to nitric and nitrous oxides via the elusive nitrite radical dianion
V. Hosseininasab, I. M DiMucci, P. Ghosh, J. A. Bertke, S. Chandrasekharan, C. J. Titus, D. Nordlund, J. H. Freed, K. M. Lancaster, T. H. Warren
Nat. Chem. 14, 1265-1269 (2022)
Supporting Information
<doi: 10.1038/s41557-022-01025-9>
PMID:
36064970
PMCID:
PMC9633411
Publication #458
|

|
|
ABSTRACT: Reduction of nitrite anions (NO2–) to nitric oxide (NO), nitrous oxide (N2O) and ultimately dinitrogen (N2) takes place in a variety of environments, including in the soil as part of the biogeochemical nitrogen cycle and in acidified nuclear waste. Nitrite reduction typically takes place within the coordination sphere of a redox-active transition metal. Here we show that Lewis acid coordination can substantially modify the reduction potential of this polyoxoanion to allow for its reduction under non-aqueous conditions (–0.74 V versus NHE). Detailed characterization confirms the formation of the borane-capped radical nitrite dianion (NO22–), which features a N(II) oxidation state. Protonation of the nitrite dianion results in the facile loss of nitric oxide (NO), whereas its reaction with NO results in disproportionation to nitrous oxide (N2O) and nitrite (NO2–). This system connects three redox levels in the global nitrogen cycle and provides fundamental insights into the conversion of NO2– to NO.
|
|
|
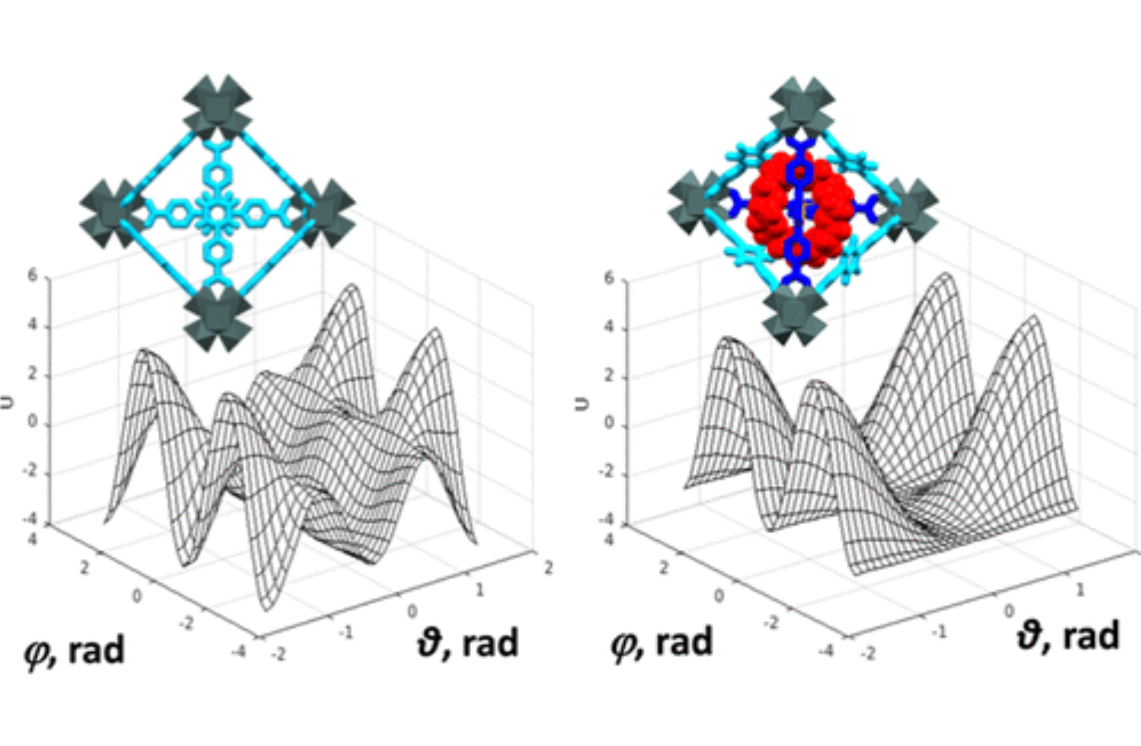
Structural Dynamics by NMR in the Solid State: II. The MOMD Perspective of the Dynamic Structure of Metal-Organic Frameworks Comprising Several Mobile Components
E. Meirovitch, Z. Liang, R. W. Schurko, S. J. Loeb, and J. H. Freed.
J. Phys. Chem. B 126, 2452-2465 (2022)
Supporting Information
<doi: 10.1021/acs.jpcb.1c10120>
PMID:
35333061
PMCID:
PMC9055879
Publication #457
|
|
|
ABSTRACT: We describe the application of the microscopic-order-macroscopic-disorder (MOMD) approach, developed for the analysis of dynamic 2H NMR lineshapes in the solid state, to unravel interactions among the constituents of metal–organic frameworks (MOFs) that comprise mobile components. MOMD was applied recently to University of Windsor Dynamic Material (UWDM) MOFs with one mobile crown ether per cavity. In this work, we study UWDM-9-d4, which comprises a mobile 2H-labeled phenyl-ring residue along with an isotopically unlabeled 24C8 crown ether. We also study UiO-68-d4, which is structurally similar to UWDM-9-d4 but lacks the crown ether. The physical picture consists of the NMR probe–the C–D bonds of the phenyl-d4 rotor–diffusing locally (diffusion tensor R) in the presence of a local ordering potential, u. For UiO-68-d4, we find it sufficient to expand u in terms of four real Wigner functions, D0|K|L, overall 2–3 kT in magnitude, with R∥ relatively fast, and R⊥; in the (2.8–5.0) × 102 s-1 range. For UWDM-9-d4, u requires only two terms 2–3 kT in magnitude and slower rate constants R∥ and R⊥. In the more crowded macrocycle-containing UWDM-9-d4 cavity, phenyl-d4 dynamics is more isotropic and is described by a simpler ordering potential. This is ascribed to cooperative phenyl-ring/macrocycle motion, which yields a dynamic structure more uniform in character. The experimental 2H spectra used here were analyzed previously with a multi-simple-mode (MSM) approach where several independent simple motional modes are combined. Where possible, similar features have been identified and used to compare the two approaches.
|
|
|
The N-Terminal Domain of Aβ40-Amyloid Fibril: The MOMD Perspective of its Dynamic Structure from NMR Lineshape Analysis
E. Meirovitch, Z. Liang, and J. H. Freed.
J. Phys. Chem. B 126, 1202-1211 (2022)
Supporting Information
<doi: 10.1021/acs.jpcb.1c10131>
PMID:
35128920
PMCID:
PMC8908910
Publication #456
|
|
|
ABSTRACT: We have developed the stochastic microscopic‐order‐macroscopic‐disorder (MOMD) approach for elucidating dynamic structures in the solid‐state from 2H NMR lineshapes. In MOMD, the probe experiences an effective/collective motional mode. The latter is described by a potential, u, which represents the local spatial‐restrictions, a local‐motional diffusion tensor, R, and key features of local geometry. Previously we applied MOMD to the well‐structured core domain of the 3‐fold‐symmetric twisted polymorph of the Aβ40‐amyloid fibril. Here, we apply it to the N‐terminal domain of this fibril. We find that the dynamic structures of the two domains are largely similar but differ in the magnitude and complexity of the key physical parameters. This interpretation differs from previous multisimple‐mode (MSM) interpretations of the same experimental data. MSM used for the two domains different combinations of simple motional modes taken to be independent. For the core domain, MOMD and MSM disagree on the character of the dynamic structure. For the N‐terminal domain, they even disagree on whether this chain segment is structurally ordered (MOMD finds that it is), and whether it undergoes a phase transition at 260 K where bulklike water located in the fibril matrix freezes (MOMD finds that it does not). These are major differences associated with an important system. While the MOMD description is a physically sound one, there are drawbacks in the MSM descriptions. The results obtained in this study promote our understanding of the dynamic structure of protein aggregates. Thus, they contribute to the effort to pharmacologically control neurodegenerative disorders believed to be caused by such aggregates.
|
|
|
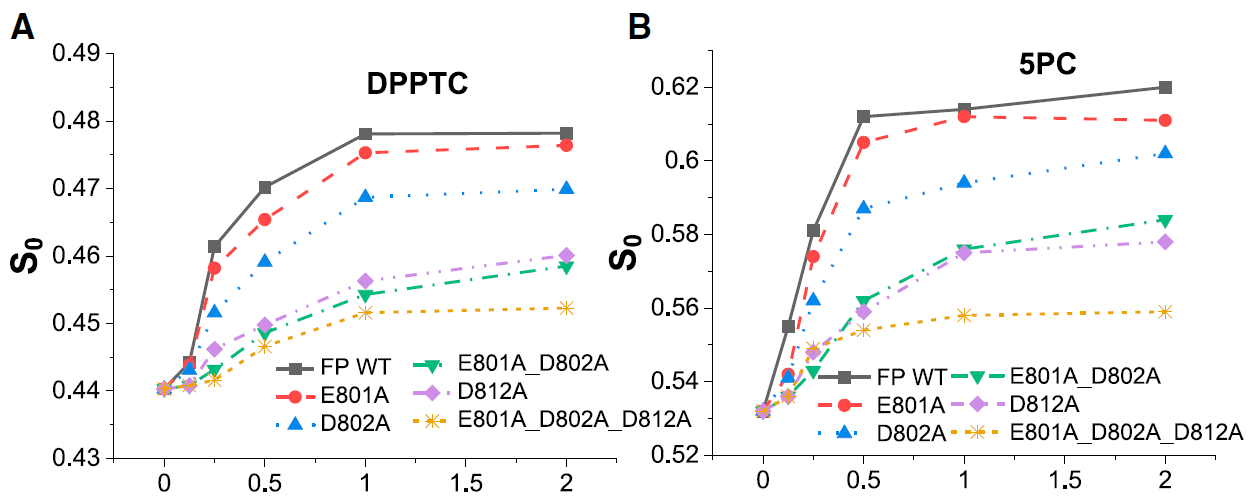
Negatively charged residues in the membrane ordering activity of SARS‐CoV‐1 and ‐2 fusion peptides
A.L. Lai and J.H. Freed
Biophys. J. 121, 207-227 (2022)
Supporting Information
<doi: 10.1016/j.bpj.2021.12.024>
PMID:
34929193
PMCID:
PMC8683214
Publication #455
|
|
|
ABSTRACT: Entry of coronaviruses into host cells is mediated by the viral spike protein. Previously, we identified the bona fide fusion peptides (FPs) for severe acute respiratory syndrome coronavirus ("SARS‐1") and severe acute respiratory syndrome coronavirus‐2 ("SARS‐2") using electron spin resonance spectroscopy. We also found that their FPs induce membrane ordering in a Ca2+‐dependent fashion. Here we study which negatively charged residues in SARS‐1 FP are involved in this binding, to build a topological model and clarify the role of Ca2+. Our systematic mutation study on the SARS‐1 FP shows that all six negatively charged residues contribute to the FP's membrane ordering activity, with D812 the dominant residue. The corresponding SARS‐2 residue D830 plays an equivalent role. We provide a topological model of how the FP binds Ca2+ ions: its two segments FP1 and FP2 each bind one Ca2+. The binding of Ca2+, the folding of FP (both studied by isothermal titration calorimetry experiments), and the ordering activity correlate very well across the mutants, suggesting that the Ca2+ helps the folding of FP in membranes to enhance the ordering activity. Using a novel pseudotyped viral particle‐liposome methodology, we monitored the membrane ordering induced by the FPs in the whole spike protein in its trimer form in real time. We found that the SARS‐1 and SARS‐2 pseudotyped viral particles also induce membrane ordering to the extent that separate FPs do, and mutations of the negatively charged residues also significantly suppress the membrane ordering activity. However, the slower kinetics of the FP ordering activity versus that of the pseudotyped viral particle suggest the need for initial trimerization of the FPs.
|
|
|
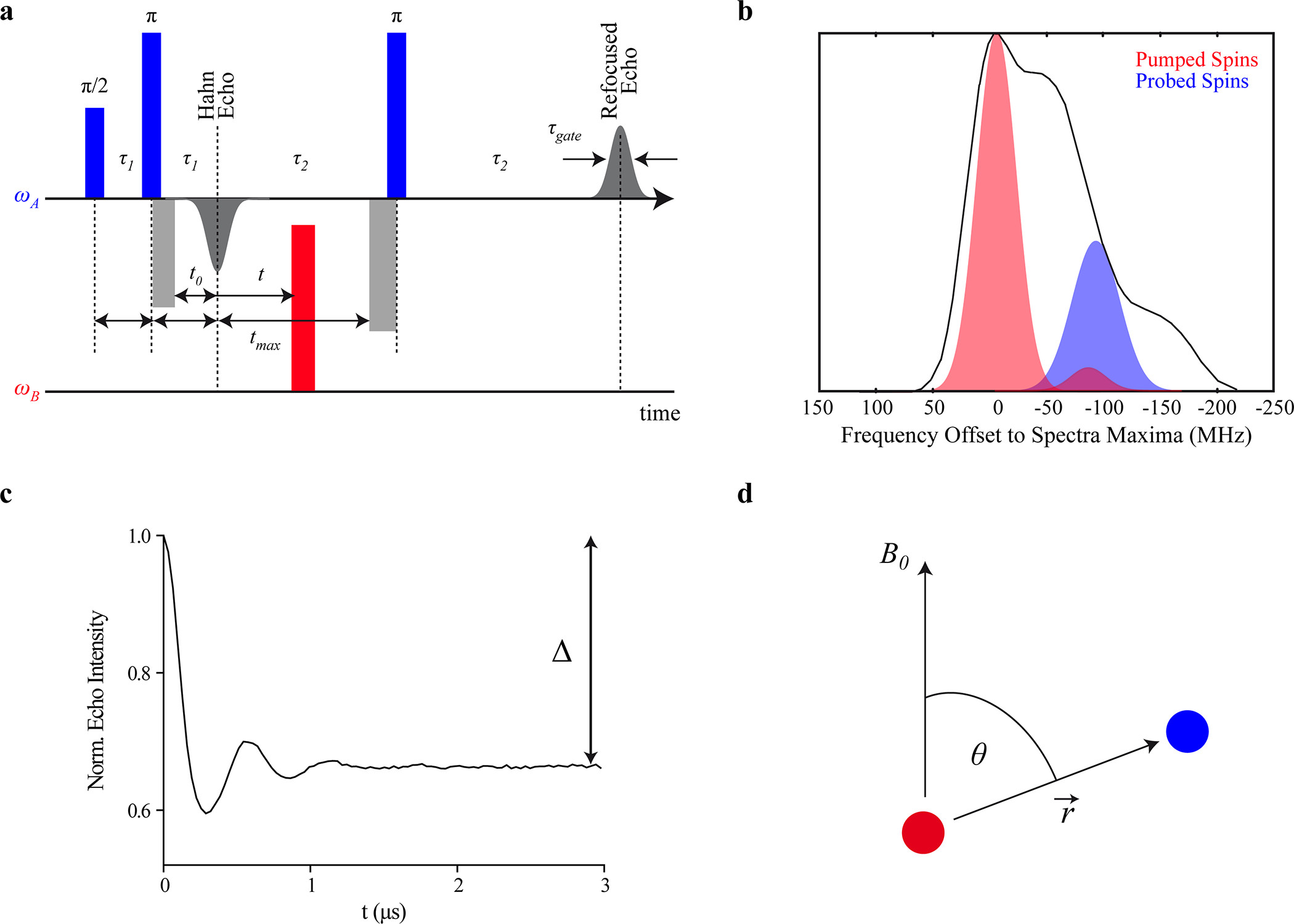
Benchmark Test and Guidelines for DEER/PELDOR Experiments on Nitroxide-Labeled Biomolecules
O. Schiemann, C. A. Heubach, D. Abdullin, K. Ackermann, M. Azarkh, E. G. Bagryanskaya, M. Drescher, B. Endeward, J. H. Freed, L. Galazzo, D. Goldfarb, T. Hett, L. Esteban Hofer, L. Fábregas Ibáñez, E. J. Hustedt, S. Kucher, I. Kuprov, J. E. Lovett, A. Meyer, S. Ruthstein, S. Saxena, S. Stoll, C. R. Timmel, M. Di Valentin, H. S. Mchaourab, T. F. Prisner, B. E. Bode, E. Bordignon, M. Bennati, and G. Jeschke.
J. Am. Chem. Soc. 143, 17875-17890 (2021)
|
|
|
Benchmark Test and Guidelines for DEER/PELDOR Experiments on Nitroxide-Labeled Biomolecules
O. Schiemann, C. A. Heubach, D. Abdullin, K. Ackermann, M. Azarkh, E. G. Bagryanskaya, M. Drescher, B. Endeward, J. H. Freed, L. Galazzo, D. Goldfarb, T. Hett, L. Esteban Hofer, L. Fábregas Ibáñez, E. J. Hustedt, S. Kucher, I. Kuprov, J. E. Lovett, A. Meyer, S. Ruthstein, S. Saxena, S. Stoll, C. R. Timmel, M. Di Valentin, H. S. Mchaourab, T. F. Prisner, B. E. Bode, E. Bordignon, M. Bennati, and G. Jeschke.
J. Am. Chem. Soc. 143, 17875-17890 (2021)
Supporting Information
<doi: 10.1021/jacs.1c07371>
PMID:
34664948
PMCID:
PMC11253894
Publication #454
|

|
|
ABSTRACT: Distance distribution information obtained by pulsed dipolar EPR spectroscopy provides an important contribution to many studies in structural biology. Increasingly, such information is used in integrative structural modeling, where it delivers unique restraints on the width of conformational ensembles. In order to ensure reliability of the structural models and of biological conclusions, we herein define quality standards for sample preparation and characterization, for measurements of distributed dipole–dipole couplings between paramagnetic labels, for conversion of the primary time‐domain data into distance distributions, for interpreting these distributions, and for reporting results. These guidelines are substantiated by a multi‐laboratory benchmark study and by analysis of data sets with known distance distribution ground truth. The study and the guidelines focus on proteins labeled with nitroxides and on double electron–electron resonance (DEER aka PELDOR) measurements and provide suggestions on how to proceed analogously in other cases.
|
|
|
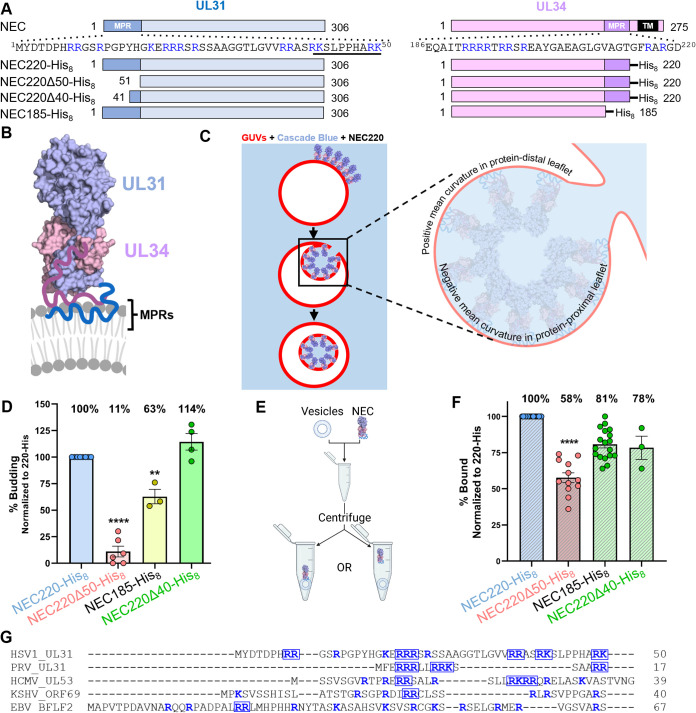
Highly Basic Clusters in the Herpes Simplex Virus 1 Nuclear Egress Complex Drive Membrane Budding by Inducing Lipid Ordering
M. K. Thorsen, A. Lai, D. P. Hoogerheide, G. C. L. Wong, J. H. Freed, and E. E. Heldwein.
mBio 12 e0154821 (2021)
Supporting Information
<doi: 10.1128/mBio.01548-21>
PMID:
34425706
PMCID:
PMC8406295
Publication #453
|
|
|
ABSTRACT: During replication of herpesviruses, capsids escape from the nucleus into the cytoplasm by budding at the inner nuclear membrane. This unusual process is mediated by the viral nuclear egress complex (NEC) that deforms the membrane around the capsid by oligomerizing into a hexagonal, membrane‐bound scaffold. Here, we found that highly basic membrane‐proximal regions (MPRs) of the NEC alter lipid order by inserting into the lipid headgroups and promote negative Gaussian curvature. We also find that the electrostatic interactions between the MPRs and the membranes are essential for membrane deformation. One of the MPRs is phosphorylated by a viral kinase during infection, and the corresponding phosphomimicking mutations block capsid nuclear egress. We show that the same phosphomimicking mutations disrupt the NEC‐membrane interactions and inhibit NEC‐mediated budding in vitro, providing a biophysical explanation for the in vivo phenomenon. Our data suggest that the NEC generates negative membrane curvature by both lipid ordering and protein scaffolding and that phosphorylation acts as an off switch that inhibits the membrane‐budding activity of the NEC to prevent capsid‐less budding.
IMPORTANCE Herpesviruses are large viruses that infect nearly all vertebrates and some invertebrates and cause lifelong infections in most of the world's population. During replication, herpesviruses export their capsids from the nucleus into the cytoplasm by an unusual mechanism in which the viral nuclear egress complex (NEC) deforms the nuclear membrane around the capsid. However, how membrane deformation is achieved is unclear. Here, we show that the NEC from herpes simplex virus 1, a prototypical herpesvirus, uses clusters of positive charges to bind membranes and order membrane lipids. Reducing the positive charge or introducing negative charges weakens the membrane deforming ability of the NEC. We propose that the virus employs electrostatics to deform nuclear membrane around the capsid and can control this process by changing the NEC charge through phosphorylation. Blocking NEC-membrane interactions could be exploited as a therapeutic strategy.
|
|
|
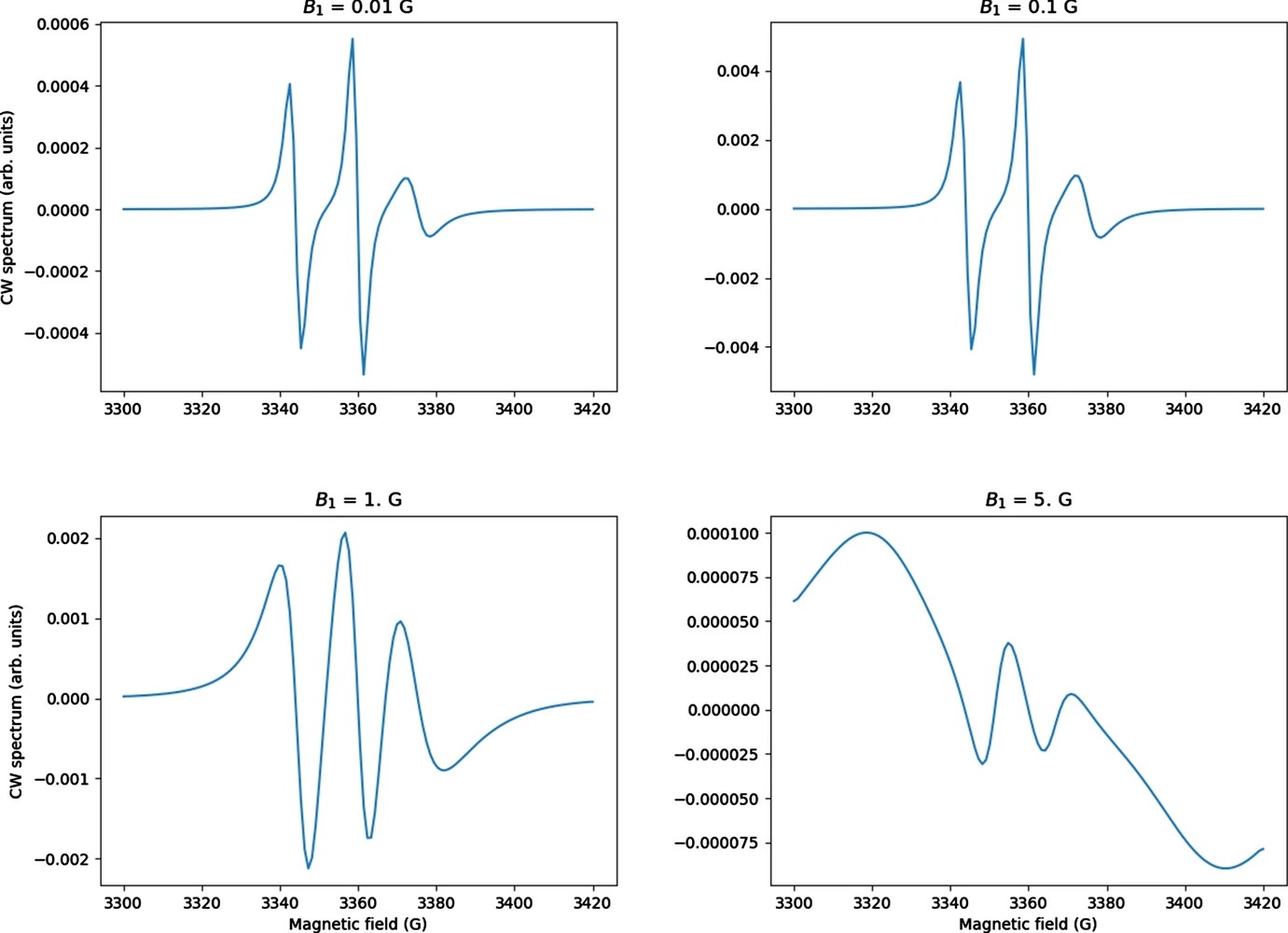
Theory and Least Squares Fitting of CW ESR Saturation Spectra Using the MOMD Model
P. Gupta, B. Dzikovski, and J. H. Freed
Appl. Magn. Reson. 53 699-715 (2021)
<doi: 10.1007/s00723-021-01390-7>
PMID:
35431460
PMCID:
PMC9012167
Publication #452
|
|
|
ABSTRACT: CW saturation experiments are widely used in ESR studies of relaxation processes in proteins and lipids. We develop the theory of saturation in ESR spectra in terms of its close relation with that of 2D‐ELDOR. Our treatment of saturation is then based on the microscopic order macroscopic disorder (MOMD) model and can be used to fit the full CW saturation spectrum, rather than fitting just the peak–peak amplitude as a function of microwave field B1 as is commonly done. This requires fewer experiments to yield effects on T1, as well as provides a more extensive dynamic structural picture, for example, for scanning experiments on different protein sites. The code is released as a publicly available software package in Python that can be used to fit CW saturation spectra from biological samples of interest.
|
|
|
Dph3 Enables Aerobic Diphthamide Biosynthesis by Donating One Iron Atom to Transform a [3Fe-4S] to a [4Fe-4S] Cluster in Dph1-Dph2
Y. Zhang, D. Su, B. Dzikovski, S. H. Majer, R. Coleman, S. Chandrasekaran, M. K. Fenwick, B. R. Crane, K. M. Lancaster, J. H. Freed, and H. Lin.
J. Am. Chem. Soc. 143, 9314-9319 (2021)
|

|
|
Dph3 Enables Aerobic Diphthamide Biosynthesis by Donating One Iron Atom to Transform a [3Fe-4S] to a [4Fe-4S] Cluster in Dph1-Dph2
Y. Zhang, D. Su, B. Dzikovski, S. H. Majer, R. Coleman, S. Chandrasekaran, M. K. Fenwick, B. R. Crane, K. M. Lancaster, J. H. Freed, and H. Lin.
J. Am. Chem. Soc. 143, 9314-9319 (2021)
Supporting Information
<doi: 10.1021/jacs.1c03956>
PMID:
34154323
PMCID:
PMC8251694
Publication #451
|

|
|
ABSTRACT: All radical S‐adenosylmethionine (radical‐SAM) enzymes, including the noncanonical radical‐SAM enzyme diphthamide biosynthetic enzyme Dph1–Dph2, require at least one [4Fe–4S](Cys)3 cluster for activity. It is well‐known in the radical‐SAM enzyme community that the [4Fe–4S](Cys)3 cluster is extremely air‐sensitive and requires strict anaerobic conditions to reconstitute activity in vitro. Thus, how such enzymes function in vivo in the presence of oxygen in aerobic organisms is an interesting question. Working on yeast Dph1–Dph2, we found that consistent with the known oxygen sensitivity, the [4Fe–4S] cluster is easily degraded into a [3Fe–4S] cluster. Remarkably, the small iron‐containing protein Dph3 donates one Fe atom to convert the [3Fe–4S] cluster in Dph1–Dph2 to a functional [4Fe–4S] cluster during the radical‐SAM enzyme catalytic cycle. This mechanism to maintain radical‐SAM enzyme activity in aerobic environments is likely general, and Dph3‐like proteins may exist to keep other radical‐SAM enzymes functional in aerobic environments.
|
|
|
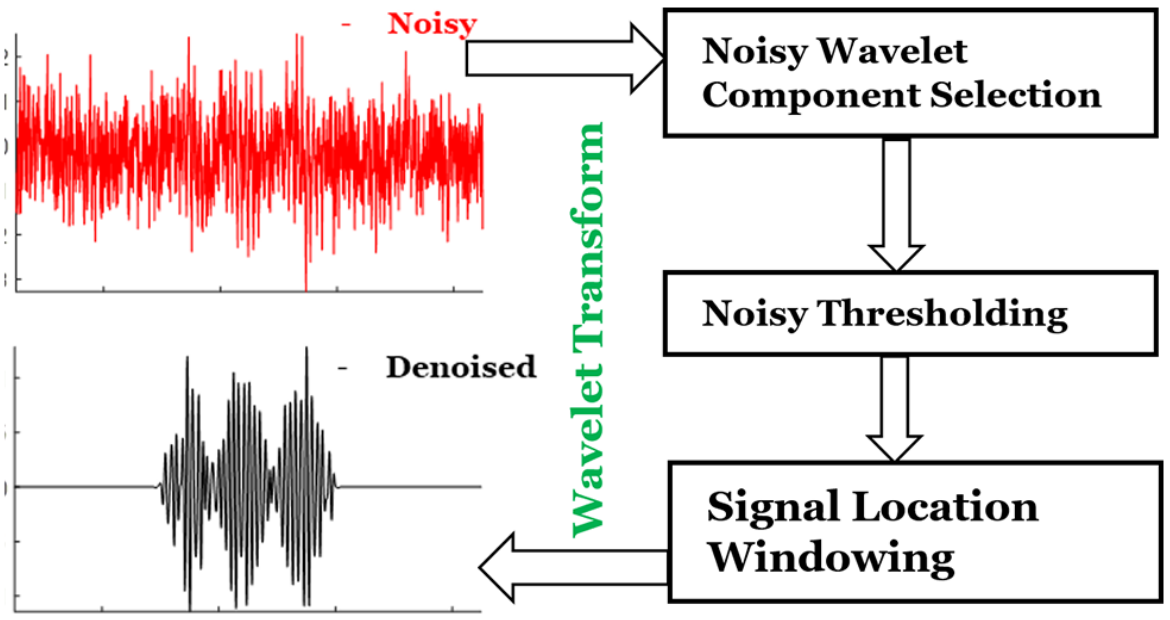
Extraction of Weak Spectroscopic Signals with High Fidelity: Examples from ESR
M. Srivastava, B. Dzikovski, and J. H. Freed.
J. Phys. Chem. A 125, 4480-4487 (2021)
Supporting Information
<doi: 10.1021/acs.jpca.1c02241>
PMID:
34009996
PMCID:
PMC8317606
Publication #450
|
|
|
ABSTRACT: Noise impedes experimental studies by reducing signal resolution and/or suppressing weak signals. Signal averaging and filtering are the primary methods used to reduce noise, but they have limited effectiveness and lack capabilities to recover signals at low signal‐to‐noise ratios (SNRs). We utilize a wavelet transform‐based approach to effectively remove noise from spectroscopic data. The wavelet denoising method we use is a significant improvement on standard wavelet denoising approaches. We demonstrate its power in extracting signals from noisy spectra on a variety of signal types ranging from hyperfine lines to overlapped peaks to weak peaks overlaid on strong ones, drawn from electron‐spin‐resonance spectroscopy. The results show that one can accurately extract details of complex spectra, including retrieval of very weak ones. It accurately recovers signals at an SNR of ˜1 and improves the SNR by about 3 orders of magnitude with high fidelity. Our examples show that one is now able to address weaker SNR signals much better than by previous methods. This new wavelet approach can be successfully applied to other spectroscopic signals.
|
|
|
SARS-CoV-2 Fusion Peptide has a Greater Membrane Perturbating Effect than SARS-CoV with Highly Specific Dependence on Ca2+
A. L. Lai and J. H. Freed.
J. Mol. Biol. 433, 166946 (2021)
<doi: 10.1016/j.jmb.2021.166946>
PMID:
33744314
PMCID:
PMC7969826
Publication #449
|

|
|
ABSTRACT: Coronaviruses are a major infectious disease threat, and include the zoonotic‐origin human pathogens SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV (SARS‐2, SARS‐1, and MERS). Entry of coronaviruses into host cells is mediated by the spike (S) protein. In our previous ESR studies, the local membrane ordering effect of the fusion peptide (FP) of various viral glycoproteins including the S of SARS‐1 and MERS has been consistently observed. We previously determined that the sequence immediately downstream from the S2′ cleavage site is the bona fide SARS‐1 FP. In this study, we used sequence alignment to identify the SARS‐2 FP, and studied its membrane ordering effect. Although there are only three residue differences, SARS‐2 FP induces even greater membrane ordering than SARS‐1 FP, possibly due to its greater hydrophobicity. This may be a reason that SARS‐2 is better able to infect host cells. In addition, the membrane binding enthalpy for SARS‐2 is greater. Both the membrane ordering of SARS‐2 and SARS‐1 FPs are dependent on Ca2+, but that of SARS‐2 shows a greater response to the presence of Ca2+. Both FPs bind two Ca2+ ions as does SARS‐1 FP, but the two Ca2+ binding sites of SARS‐2 exhibit greater cooperativity. This Ca2+ dependence by the SARS‐2 FP is very ion‐specific. These results show that Ca2+ is an important regulator that interacts with the SARS‐2 FP and thus plays a significant role in SARS‐2 viral entry. This could lead to therapeutic solutions that either target the FP‐calcium interaction or block the Ca2+ channel.
|
|
|
Microsecond dynamics in proteins by two-dimensional ESR. II. Addressing computational challenges
P. Gupta, K. Chaudhari, and J. H. Freed.
J. Chem. Phys. 154, 084115 (2021)
Supporting Information
<doi: 10.1063/5.0042441>
PMID:
33639766
PMCID:
PMC7928224
Publication #448
|
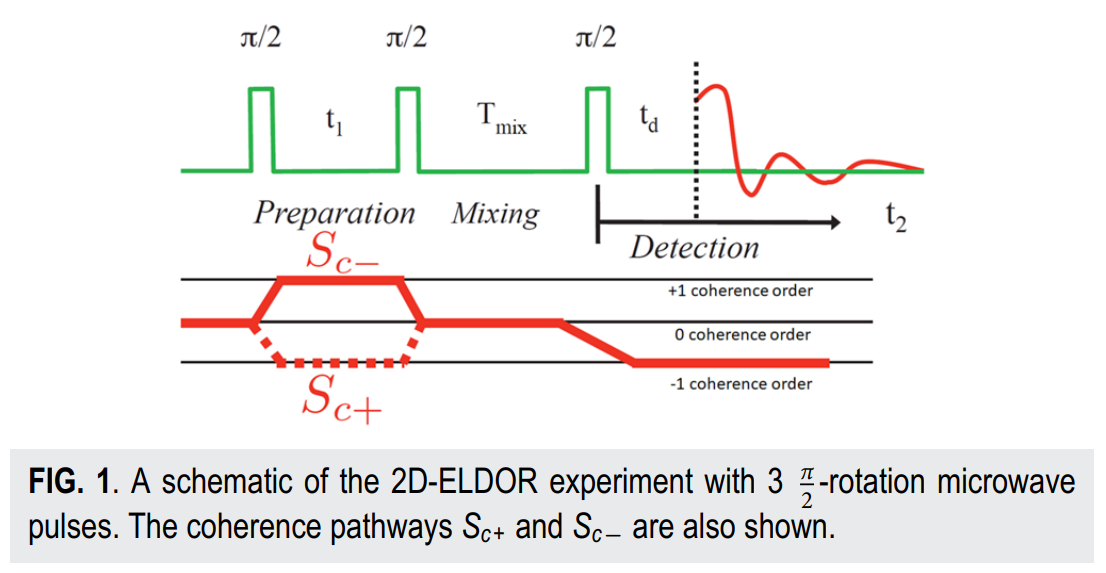
|
|
ABSTRACT: Two‐dimensional electron‐electron double resonance (2D‐ELDOR) provides extensive insight into molecular motions. Recent developments permitting experiments at higher frequencies (95 GHz) provide molecular orientational resolution, enabling a clearer description of the nature of the motions. In previous work, we provided simulations for the case of domain motions within proteins that are themselves slowly tumbling in a solution. In order to perform these simulations, it was found that the standard approach of solving the relevant stochastic Liouville equation using the efficient Lanczos algorithm for this case breaks down, so algorithms were employed that rely on the Arnoldi iteration. While they lead to accurate simulations, they are very time‐consuming. In this work, we focus on a variant known as the rational Arnoldi algorithm. We show that this can achieve a significant reduction in computation time. The stochastic Liouville matrix, which is of very large dimension, N, is first reduced to a much smaller dimension, m, e.g., from N ˜ O(104) to m ˜ 60, that spans the relevant Krylov subspace from which the spectrum is predicted. This requires the selection of the m frequency shifts to be utilized. A method of adaptive shift choice is introduced to optimize this selection. We also find that these procedures help in optimizing the pruning procedure that greatly reduces the dimension of the initial N dimensional stochastic Liouville matrix in such subsequent computations.
|
|
|
Microsecond Exchange Processes Studied by Two‐Dimensional ESR at 95 GHz
B. Dzikovski, V. V. Khramtsov, S. Chandrasekaran, C. Dunnam, M. Shah, and J. H. Freed.
J. Am. Chem. Soc. 142, 21368-21381 (2020)
Supporting Information
<doi: 10.1021/jacs.0c09469>
PMID:
33305945
PMCID:
PMC7810061
Publication #447
|
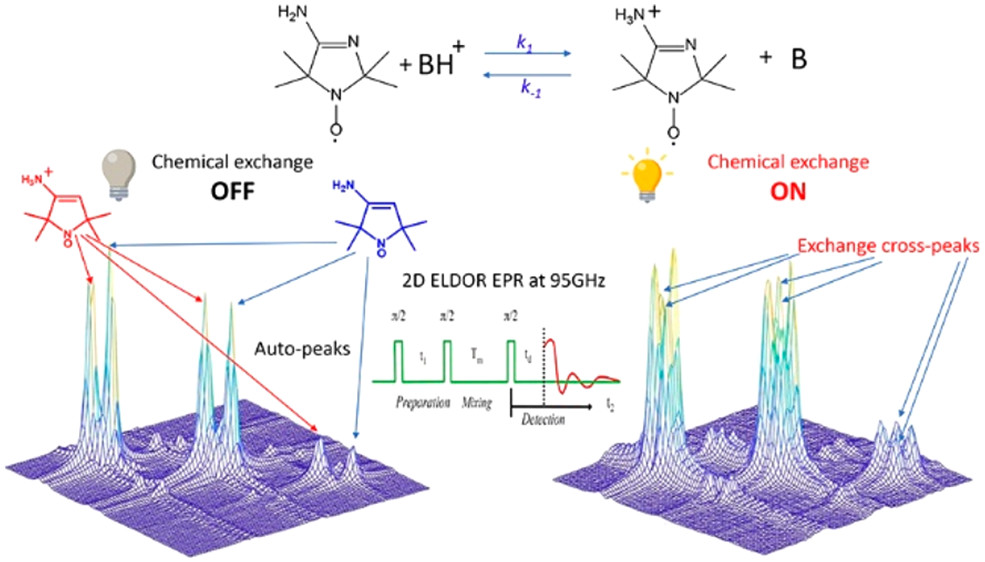
|
|
ABSTRACT: Exchange processes which include conformational change, protonation/deprotonation, and binding equilibria are routinely studied by 2D exchange NMR techniques, where information about the exchange of nuclei between environments with different NMR shifts is obtained from the development of cross‐peaks. Whereas 2D NMR enables the real time study of millisecond and slower exchange processes, 2D ESR in the form of 2D‐ELDOR (two‐dimensional electron‐electron double resonance) has the potential for such studies over the nanosecond to microsecond real time scales. Cross‐peak development due to chemical exchange has been seen previously for semiquinones in ESR, but this is not possible for most common ESR probes, such as nitroxides, studied at typical ESR frequencies because, unlike NMR, the exchanging states yield ESR signals that are not resolved from each other within their respective line widths. But at 95 GHz, it becomes possible to resolve them in many cases because of the increased g‐factor resolution. The 95 GHz instrumental developments occurring at ACERT now enable such studies. We demonstrate these new capabilities in two studies: (A) the protonation/deprotonation process for a pH‐sensitive imidazoline spin label in aqueous solution where the exchange rate and the population ratio of the exchanging states are controlled by the concentration and pH of the buffer solution, respectively, and (B) a nitroxide radical partitioning between polar (aqueous) and nonpolar (phospholipid) environments in multilamellar lipid vesicles, where the cross‐peak development arises from the exchange of the nitroxide between the two phases. This work represents the first example of the observation and analysis of cross‐peaks arising from chemical exchange processes involving nitroxide spin labels.
|
|
|
Engineered chemotaxis core signaling units indicate a constrained kinase-off state
A. R. Muok, T. K. Chua, M. Srivastava, W. Yang, Z. Maschmann, P. P. Borbat, J. Chong, S. Zhang, J. H. Freed, A. Briegel, B. R. Crane.
Sci. Signal. 13, eabc1328 (2020)
|
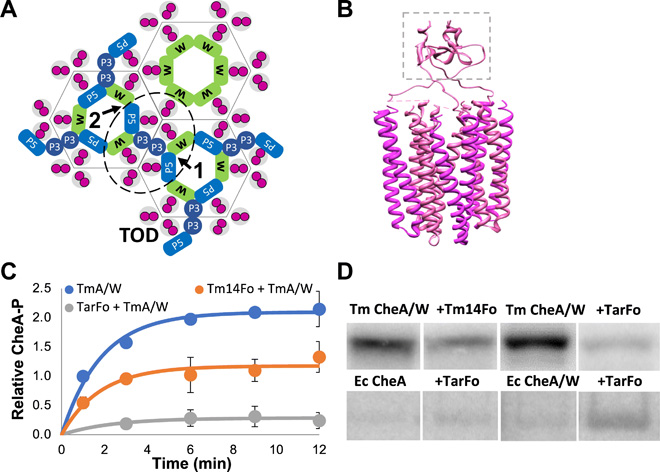
|
|
Engineered chemotaxis core signaling units indicate a constrained kinase-off state
A. R. Muok, T. K. Chua, M. Srivastava, W. Yang, Z. Maschmann, P. P. Borbat, J. Chong, S. Zhang, J. H. Freed, A. Briegel, B. R. Crane.
Sci. Signal. 13, eabc1328 (2020)
Supporting Information
<doi: 10.1126/scisignal.abc1328>
PMID:
33172954
PMCID:
PMC7790435
Publication #446
|

|
|
ABSTRACT: Bacterial chemoreceptors, the histidine kinase CheA, and the coupling protein CheW form transmembrane molecular arrays with remarkable sensing properties. The receptors inhibit or stimulate CheA kinase activity depending on the presence of attractants or repellants, respectively. We engineered chemoreceptor cytoplasmic regions to assume a trimer of receptor dimers configuration that formed well‐defined complexes with CheA and CheW and promoted a CheA kinase‐off state. These mimics of core signaling units were assembled to homogeneity and investigated by site‐directed spin‐labeling with pulse‐dipolar electron‐spin resonance spectroscopy (PDS), small‐angle x‐ray scattering, targeted protein cross‐linking, and cryo–electron microscopy. The kinase‐off state was especially stable, had relatively low domain mobility, and associated the histidine substrate and docking domains with the kinase core, thus preventing catalytic activity. Together, these data provide an experimentally restrained model for the inhibited state of the core signaling unit and suggest that chemoreceptors indirectly sequester the kinase and substrate domains to limit histidine autophosphorylation.
|
|
|
Structural Dynamics by NMR in the Solid State: The Unified MOMD Perspective Applied to Organic Frameworks with Interlocked Molecules
E. Meirovitch, Z. Liang, and J. H. Freed.
J. Phys. Chem. B 124, 6225-6235 (2020)
Supporting Information
<doi: 10.1021/acs.jpcb.0c03687>
PMID:
32584038
PMCID:
PMC7666760
Publication #445
|
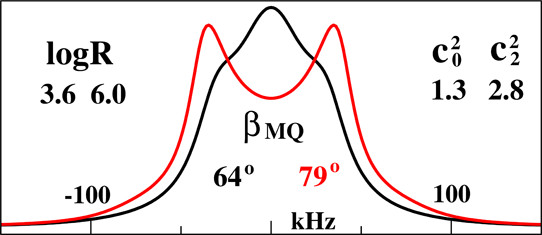
|
|
ABSTRACT: The microscopic-order-macroscopic-disorder (MOMD) approach for NMR lineshape analysis has been applied to the University of Windsor Dynamic Materials (UWDM) of types 1, 2, α-3, β-3, and 5, which are metal–organic frameworks (MOFs) comprising mobile mechanically interlocked molecules (MIMs). The mobile MIM components are selectively deuterated crown ether macrocycles – 24C6, 22C6, and B24C6. Their motion is described in MOMD by an effective/collective dynamic mode characterized by a diffusion tensor, R, a restricting/ordering potential, u, expanded in the Wigner rotation matrix elements, D0,KL, and features of local geometry. Experimental 2H lineshapes are available over 220 K (on average) and in some cases 320 K. They are reproduced with axial R, u given by the terms D0,02 and D0,|2|2, and established local geometry. For UWDM of types 1, β-3, and 5, where the macrocycle resides in a relatively loose space, u is in the 1–3 kT, R∥ in the (1.0–2.5) × 106 s–1, and R⊥ in the (0.4–2.5) × 104 s–1 range; the deuterium atom is bonded to a carbon atom with tetrahedral coordination character. For UWDM of types 2 and α-3, where the macrocycle resides in a much tighter space, a substantial change in the symmetry of u and the coordination character of the 2H-bonded carbon are detected at higher temperatures. The activation energies for R∥ and R⊥ are characteristic of each system. The MOMD model is general; effective/collective dynamic modes are treated. The characteristics of motion, ordering, and geometry are physically well-defined; they differ from case to case in extent and symmetry but not in essence. Physical clarity and consistency provide new insights. A previous interpretation of the same experimental data used models consisting of collections of independent simple motions. These models are specific to each case and temperature. Within their scope, generating consistent physical pictures and comparing cases are difficult; possible collective modes are neglected.
|
|
|
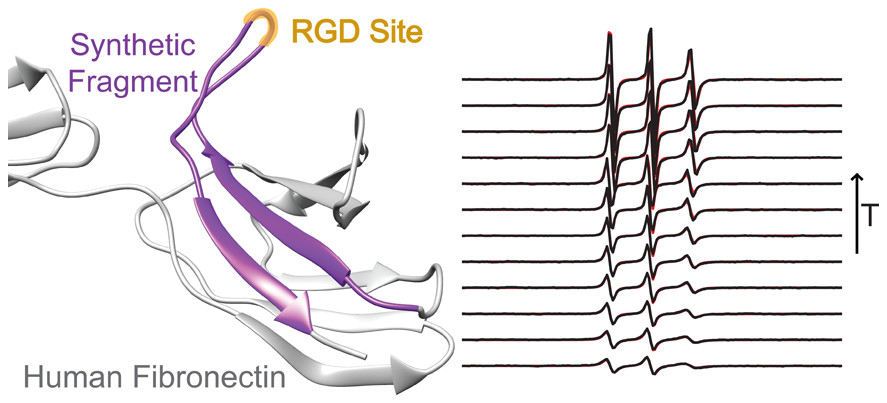
Conformational Dynamics in Extended RGD-Containing Peptides
W. R. Lindemann, A. J. Mijalis, J. L. Alonso, P. P. Borbat, J. H. Freed, M. A. Arnaout, B. L. Pentelute, and J. H. Ortony.
Biomacromolecules 21, 2786-2794 (2020)
Supporting Information
<doi: 10.1021/acs.biomac.0c00506>
PMID:
32469507
PMCID:
PMC7388056
Publication #444
|
|
|
ABSTRACT: RGD is a prolific example of a tripeptide used in biomaterials for cell adhesion, but the potency of free or surface-bound RGD tripeptide is orders-of-magnitude less than the RGD domain within natural proteins. We designed a set of peptides with varying lengths, composed of fragments of fibronectin protein whose central three residues are RGD, in order to vary their conformational behavior without changing the binding site's chemical environment. With these peptides, we measure the conformational dynamics and transient structure of the active site. Our studies reveal how flanking residues affect conformational behavior and integrin binding. We find that disorder of the binding site is important to the potency of RGD peptides and that transient hydrogen bonding near the RGD site affects both the energy landscape roughness of the peptides and peptide binding. This phenomenon is independent of longer-range folding interactions and helps explain why short binding sequences, including RGD itself, do not fully replicate the integrin-targeting properties of extracellular matrix proteins. Our studies reinforce that peptide binding is a holistic event and fragments larger than those directly involved in binding should be considered in the design of peptide epitopes for functional biomaterials.
|
|
|
Microsecond dynamics in proteins by two-dimensional ESR: Predictions
P. Gupta, Z. Liang, and J. H. Freed.
J. Chem. Phys. 152, 214112 (2020)
Supporting Information
<doi: 10.1063/5.0008094>
PMID:
32505151
PMCID:
PMC7863697
Publication #443
|

|
|
ABSTRACT: Two-dimensional electron-electron double resonance (2D-ELDOR) provides extensive insight into molecular motions. Recent developments permitting experiments at higher frequencies (95 GHz) provide molecular orientational resolution, enabling a clearer description of the nature of the motions. In this work, simulations are provided for the example of domain motions within proteins that are themselves slowly tumbling in solution. These show the nature of the exchange cross-peaks that are predicted to develop in real time from such domain motions. However, we find that the existing theoretical methods for computing 2D-ELDOR experiments over a wide motional range begin to fail seriously when applied to very slow motions characteristic of proteins in solution. One reason is the failure to obtain accurate eigenvectors and eigenvalues of the complex symmetric stochastic Liouville matrices describing the experiment when computed by the efficient Lanczos algorithm in the range of very slow motion. Another, perhaps more serious, issue is that these matrices are "non-normal," such that for the very slow motional range even rigorous diagonalization algorithms do not yield the correct eigenvalues and eigenvectors. We have employed algorithms that overcome both these issues and lead to valid 2D-ELDOR predictions even for motions approaching the rigid limit. They are utilized to describe the development of cross-peaks in 2D-ELDOR at 95 GHz for a particular case of domain motion.
|
|
|
George K. Fraenkel, Electron Spin Resonance Pioneer
J.H. Freed.
In Pioneers of Magnetic Resonance. Strom, E. T., Mainz, V. V., Eds. American Chemical Society: Washington, DC, ACS Symposium Series, 2020; Volume 1349, Chapter 8, pp. 137-154.
|
|
|
George K. Fraenkel, Electron Spin Resonance Pioneer
J.H. Freed.
In Pioneers of Magnetic Resonance. Strom, E. T., Mainz, V. V., Eds. American Chemical Society: Washington, DC, ACS Symposium Series, 2020; Volume 1349, Chapter 8, pp. 137-154.
<doi: 10.1021/bk-2020-1349.ch008>
PMID: [None-book] PMCID:
[None-book]
Publication #442
|
|
|
ABSTRACT: George K. Fraenkel (1921-2009), although less known today, was one of the leading pioneers in the development and use of Electron Spin Resonance (ESR) techniques for studying the structure and dynamical interactions of molecules. Together with his students he developed high-sensitivity, high-resolution spectrometers that enabled them to pioneer the study of ESR in free radicals in solution. His work provided breakthroughs that led to advances in several fields of chemistry, and laid the foundations for later research on the properties of biological systems. This chapter provides a thematic overview, arranged chronologically, of his various studies. Short descriptions are provided of his achievements in these areas, based on his most important papers, so that present-day researchers can appreciate the extent of his accomplishments.
|
|
|
High-yield production in E. coli and characterization of full-length functional p13II protein from human T-cell leukemia virus type 1
E. R. Georgieva, P. P. Borbat, C. Fanouraki, J. H. Freed.
Protein Expr. Purif. 173, 105659 (2020)
Supporting Information
<doi: 10.1016/j.pep.2020.105659>
PMID:
32360379
PMCID:
PMC7266171
Publication #441
|
|
|
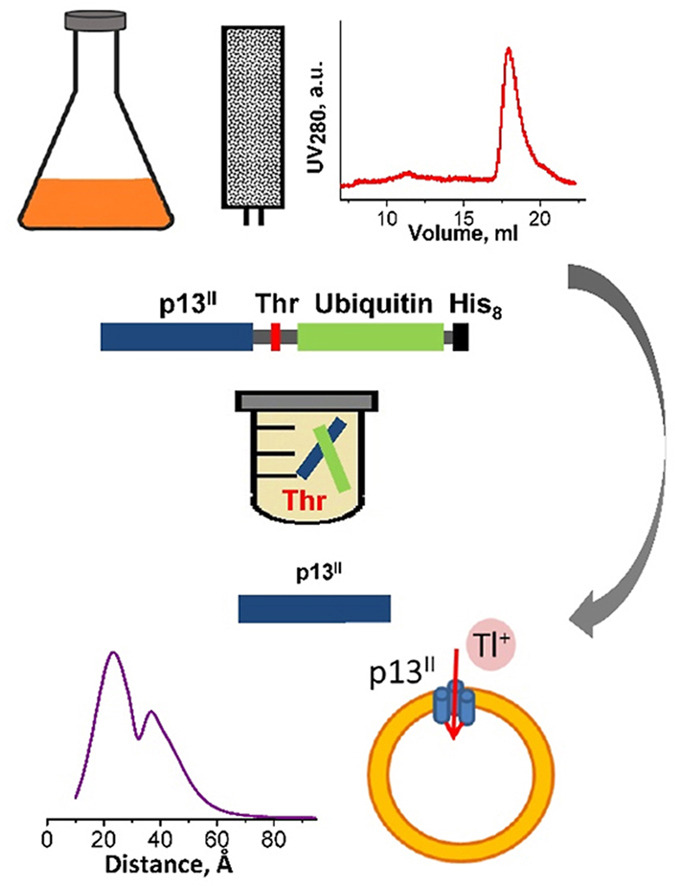
ABSTRACT: Human T-cell leukemia virus type 1 is an oncovirus that causes aggressive adult T-cell leukemia but is also responsible for severe neurodegenerative and endocrine disorders. Combatting HTLV-1 infections requires a detailed understanding of the viral mechanisms in the host. Therefore, in vitro studies of important virus-encoded proteins would be critical. Our focus herein is on the HTLV-1-encoded regulatory protein p13II, which interacts with the inner mitochondrial membrane, increasing its permeability to cations (predominantly potassium, K+). Thereby, this protein affects mitochondrial homeostasis. We report on our progress in developing specific protocols for heterologous expression of p13II in E. coli, and methods for its purification and characterization. We succeeded in producing large quantities of highly-pure full-length p13II, deemed to be its fully functional form. Importantly, our particular approach based on the fusion of ubiquitin to the p13II C-terminus was instrumental in increasing the persistently low expression of soluble p13II in its native form. We subsequently developed approaches for protein spin labeling and a conformation study using double electron-electron resonance (DEER) spectroscopy and a fluorescence-based cation uptake assay for p13II in liposomes. Our DEER results point to large protein conformation changes occurring upon transition from the soluble to the membrane-bound state. The functional assay on p13II-assisted transport of thallium (Tl+) through the membrane, wherein Tl+ substituted for K+, suggests transmembrane potential involvement in p13II function. Our study lays the foundation for expansion of in vitro functional and structural investigations on p13II and would aid in the development of structure-based protein inhibitors and markers.
|
|
|
Ca2+ Ions Promote Fusion of Middle East Respiratory Syndrome Coronavirus with Host Cells and Increase Infectivity
M. R. Straus, T. Tang, A. L. Lai, A. Flegel, M. Bidon, J. H. Freed, S. Daniel, G. R. Whittaker.
J. Virol. 94, e00426-20 (2020)
Supporting Information
<doi: 10.1128/JVI.00426-20>
PMID:
32295925
PMCID:
PMC7307142
Publication #440
|
|
|
ABSTRACT: Fusion with, and subsequent entry into, the host cell is one of the critical steps in the life cycle of enveloped viruses. For Middle East respiratory syndrome coronavirus (MERS-CoV), the spike (S) protein is the main determinant of viral entry. Proteolytic cleavage of the S protein exposes its fusion peptide (FP), which initiates the process of membrane fusion. Previous studies on the related severe acute respiratory syndrome coronavirus (SARS-CoV) FP have shown that calcium ions (Ca2+) play an important role in fusogenic activity via a Ca2+ binding pocket with conserved glutamic acid (E) and aspartic acid (D) residues. SARS-CoV and MERS-CoV FPs share a high sequence homology, and here, we investigated whether Ca2+ is required for MERS-CoV fusion by screening a mutant array in which E and D residues in the MERS-CoV FP were substituted with neutrally charged alanines (A). Upon verifying mutant cell surface expression and proteolytic cleavage, we tested their ability to mediate pseudoparticle (PP) infection of host cells in modulating Ca2+ environments. Our results demonstrate that intracellular Ca2+ enhances MERS-CoV wild-type (WT) PP infection by approximately 2-fold and that E891 is a crucial residue for Ca2+ interaction. Subsequent electron spin resonance (ESR) experiments revealed that this enhancement could be attributed to Ca2+ increasing MERS-CoV FP fusion-relevant membrane ordering. Intriguingly, isothermal calorimetry showed an approximate 1:1 MERS-CoV FP to Ca2+ ratio, as opposed to an 1:2 SARS-CoV FP to Ca2+ ratio, suggesting significant differences in FP Ca2+ interactions of MERS-CoV and SARS-CoV FP despite their high sequence similarity.
IMPORTANCE Middle East respiratory syndrome coronavirus (MERS-CoV) is a major emerging infectious disease with zoonotic potential and has reservoirs in dromedary camels and bats. Since its first outbreak in 2012, the virus has repeatedly transmitted from camels to humans, with 2,468 confirmed cases causing 851 deaths. To date, there are no efficacious drugs and vaccines against MERS-CoV, increasing its potential to cause a public health emergency. In order to develop novel drugs and vaccines, it is important to understand the molecular mechanisms that enable the virus to infect host cells. Our data have found that calcium is an important regulator of viral fusion by interacting with negatively charged residues in the MERS-CoV FP region. This information can guide therapeutic solutions to block this calcium interaction and also repurpose already approved drugs for this use for a fast response to MERS-CoV outbreaks.
|
|
|
Calcium Ions Directly Interact with the Ebola Virus Fusion Peptide To Promote Structure–Function Changes That Enhance Infection
L. Nathan, A. L. Lai, J. K. Millet, M. R. Straus, J. H. Freed, G. R. Whittaker, and S. Daniel.
ACS Infect. Dis. 6, 250-260 (2020)
Supporting Information
<doi: 10.1021/acsinfecdis.9b00296>
PMID:
31746195
PMCID:
PMC7040957
Publication #439
|
|
|
ABSTRACT: Ebola virus disease is a serious global health concern given its periodic occurrence, high lethality, and the lack of approved therapeutics. Certain drugs that alter intracellular calcium, particularly in endolysosomes, have been shown to inhibit Ebola virus infection; however, the underlying mechanism is unknown. Here, we provide evidence that Zaire ebolavirus (EBOV) infection is promoted in the presence of calcium as a result of the direct interaction of calcium with the EBOV fusion peptide (FP). We identify the glycoprotein residues D522 and E540 in the FP as functionally critical to EBOV's interaction with calcium. We show using spectroscopic and biophysical assays that interactions of the fusion peptide with Ca2+directly targets the Ebola virus fusion peptide and influences its conformation. As these residues are highly conserved across the Filoviridae, calcium's impact on fusion, and subsequently infectivity, is a key interaction that can be leveraged for developing strategies to defend against Ebola infection. This mechanistic insight provides a rationale for the use of calcium-interfering drugs already approved by the FDA as therapeutics against Ebola and enables further development of novel drugs to combat the virus.
|
|
|
The asymmetric function of Dph1–Dph2 heterodimer in diphthamide biosynthesis
M. Dong, E. E. Dando, I. Kotliar, X. Su, B. Dzikovski, J. H. Freed, and H. Lin.
J. Biol. Inorg. Chem. 24, 777-782 (2019)
<doi: 10.1007/s00775-019-01702-0>
PMID:
31463593
PMCID:
PMC6893874
Publication #438
|

|
|
ABSTRACT: Diphthamide, the target of diphtheria toxin, is a post-translationally modified histidine residue found in archaeal and eukaryotic translation elongation factor 2 (EF2). In the first step of diphthamide biosynthesis, a [4Fe–4S] cluster-containing radical SAM enzyme, Dph1–Dph2 heterodimer in eukaryotes or Dph2 homodimer in archaea, cleaves S-adenosylmethionine and transfers the 3-amino-3-carboxypropyl group to EF2. It was demonstrated previously that for the archaeal Dph2 homodimer, only one [4Fe–4S] cluster is necessary for the in vitro activity. Here, we demonstrate that for the eukaryotic Dph1–Dph2 heterodimer, the [4Fe–4S] cluster-binding cysteine residues in each subunit are required for diphthamide biosynthesis to occur in vivo. Furthermore, our in vitro reconstitution experiments with Dph1–Dph2 mutants suggested that the Dph1 cluster serves a catalytic role, while the Dph2 cluster facilitates the reduction of the Dph1 cluster by the physiological reducing system Dph3/Cbr1/NADH. Our results reveal the asymmetric functional roles of the Dph1–Dph2 heterodimer and may help to understand how the Fe–S clusters in radical SAM enzymes are reduced in biology.
|
|
|
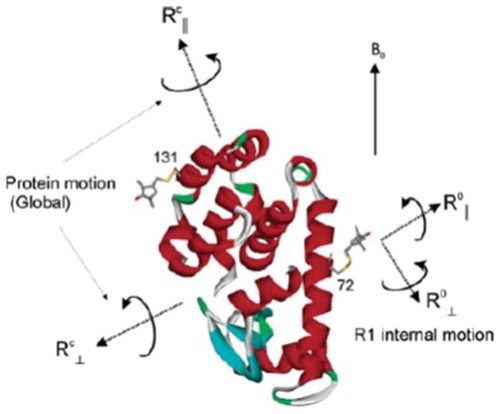
Local ordering and dynamics in anisotropic media by magnetic resonance: from liquid crystals to proteins
E. Meirovitch & J. H. Freed.
Liq. Cryst. 47, 1926-1954 (2020)
<doi: 10.1080/02678292.2019.1622158>
PMID:
32435078
PMCID:
PMC7239324
Publication #437
|
|
|
ABSTRACT: Magnetic resonance methods have been used extensively for over 50 years to elucidate molecular structure and dynamics of liquid crystals (LCs), providing information quite unique in its rigour and extent. The ESR- or NMR-active probe is often a solute molecule reporting on characteristics associated with the surrounding (LC) medium, which exerts the spatial restrictions on the probe. The theoretical approaches developed for LCs are applicable to anisotropic media in general. Of particular interest is the interior space of a globular protein labelled, e.g. with a nitroxide moiety or a 15N–1H bond. The ESR or NMR label plays the role of the probe and the internal protein surroundings the role of the anisotropic medium. A general feature of the restricted motions is the local ordering, i.e. the nature, magnitude and symmetry of the spatial restraints exerted at the site of the moving probe. This property is the main theme of the present review article. We outline its treatment in our work from both the theoretical and the experimental points of view, highlighting the new physical insights gained. Our illustrations include studies on thermotropic (nematic and smectic) and lyotropic liquid crystals formed by phospholipids, in addition to studies of proteins.
|
|
|
Insights into histidine kinase activation mechanisms from the monomeric blue light sensor EL346
I. Dikiy, U. R. Edupuganti, R. R. Abzalimov, P. P. Borbat, M. Srivastava, J. H. Freed, and K. H. Gardner.
Proc. Natl. Acad. Sci. 116, 4963-4972 (2019)
Supporting Information
<doi: 10.1073/pnas.1813586116>
PMID:
30808807
PMCID:
PMC6421462
Publication #436
|

|
|
SIGNIFICANCE: All living things must sense and react to their environment. Many single-celled organisms do so by using two-component systems, most simply consisting of a sensor histidine kinase and a response regulator. These systems are involved in pathogenicity pathways and can be targeted by new antibiotics. However, the molecular mechanisms used by histidine kinases to translate sensing into responses are not well understood. To probe this general question, we apply a combination of biophysical techniques to a monomeric histidine kinase that senses blue light to determine the structural changes occurring upon activation. We find these changes to be similar to those predicted for the common dimeric histidine kinases, illustrating that the mechanism of activation is conserved regardless of oligomeric state.
ABSTRACT: Translation of environmental cues into cellular behavior is a necessary process in all forms of life. In bacteria, this process frequently involves two-component systems in which a sensor histidine kinase (HK) autophosphorylates in response to a stimulus before subsequently transferring the phosphoryl group to a response regulator that controls downstream effectors. Many details of the molecular mechanisms of HK activation are still unclear due to complications associated with the multiple signaling states of these large, multidomain proteins. To address these challenges, we combined complementary solution biophysical approaches to examine the conformational changes upon activation of a minimal, blue-light–sensing histidine kinase from Erythrobacter litoralis HTCC2594, EL346. Our data show that multiple conformations coexist in the dark state of EL346 in solution, which may explain the enzyme's residual dark-state activity. We also observe that activation involves destabilization of the helices in the dimerization and histidine phosphotransfer-like domain, where the phosphoacceptor histidine resides, and their interactions with the catalytic domain. Similar light-induced changes occur to some extent even in constitutively active or inactive mutants, showing that light sensing can be decoupled from activation of kinase activity. These structural changes mirror those inferred by comparing X-ray crystal structures of inactive and active HK fragments, suggesting that they are at the core of conformational changes leading to HK activation. More broadly, our findings uncover surprising complexity in this simple system and allow us to outline a mechanism of the multiple steps of HK activation.
|
|
|
|
|
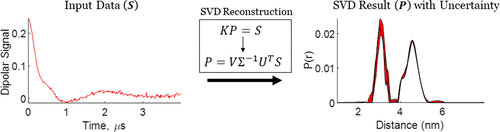
Singular Value Decomposition Method To Determine Distance Distributions in Pulsed Dipolar Electron Spin Resonance: II. Estimating Uncertainty
M. Srivastava and J. H. Freed.
J. Phys. Chem. A 123, 359-370 (2019)
Supporting Information
<doi: 10.1021/acs.jpca.8b07673>
PMID:
30525624
PMCID:
PMC6372347
Publication #434
|
|
|
ABSTRACT: This paper is a continuation of the method introduced by Srivastava and Freed (2017) that is a new method based on truncated singular value decomposition (TSVD) for obtaining physical results from experimental signals without any need for Tikhonov regularization or other similar methods that require a regularization parameter. We show here how to estimate the uncertainty in the SVD-generated solutions. The uncertainty in the solution may be obtained by finding the minimum and maximum values over which the solution remains converged. These are obtained from the optimum range of singular value contributions, where the width of this region depends on the solution point location (e.g., distance) and the signal-to-noise ratio (SNR) of the signal. The uncertainty levels typically found are very small with substantial SNR of the (denoised) signal, emphasizing the reliability of the method. With poorer SNR, the method is still satisfactory but with greater uncertainty, as expected. Pulsed dipolar electron spin resonance spectroscopy experiments are used as an example, but this TSVD approach is general and thus applicable to any similar experimental method wherein singular matrix inversion is needed to obtain the physically relevant result. We show that the Srivastava–Freed TSVD method along with the estimate of uncertainty can be effectively applied to pulsed dipolar electron spin resonance signals with SNR > 30, and even for a weak signal (e.g., SNR ≈ 3) reliable results are obtained by this method, provided the signal is first denoised using wavelet transforms (WavPDS).
|
|
|
Cofactors are essential constituents of stable and seeding-active tau fibrils
Y. Fichou, Y. Lin, J. N. Rauch, M. Vigers, Z. Zeng, M. Srivastava, T. J. Keller, J. H. Freed, K. S. Kosik, and S. Han.
Proc. Natl. Acad. Sci. 115, 13234-13239 (2018)
|

|
|
Cofactors are essential constituents of stable and seeding-active tau fibrils
Y. Fichou, Y. Lin, J. N. Rauch, M. Vigers, Z. Zeng, M. Srivastava, T. J. Keller, J. H. Freed, K. S. Kosik, and S. Han.
Proc. Natl. Acad. Sci. 115, 13234-13239 (2018)
Supporting Information
<doi: 10.1073/pnas.1810058115>
PMID:
30538196
PMCID:
PMC6310788
Publication #433
|

|
|
SIGNIFICANCE: The tau protein is involved in Alzheimer's and other neurodegenerative diseases, where the location, morphology, and quantity of amyloid fibrils composed of tau correlate with the disease type and stage. While tau fibrillary aggregates have been colocalized in brains together with several cofactors, their role in fibril formation, structure, and seeding has been largely neglected. We show that seeding of tau aggregation is facilitated by polyanionic cofactors, and that seeded or recombinant mature fibrils depolymerize into monomers when their cofactor is removed. We show that cofactor-assisted seeding with mouse brain-derived tau fibrils yielded tau fibrils with distinct and narrowed structural properties compared with heparin-induced fibrils, suggesting that the fibrillar templates tuned the structure of the seeded fibril.
ABSTRACT: Amyloid fibrils are cross-β–rich aggregates that are exceptionally stable forms of protein assembly. Accumulation of tau amyloid fibrils is involved in many neurodegenerative diseases, including Alzheimer's disease (AD). Heparin-induced aggregates have been widely used and assumed to be a good tau amyloid fibril model for most biophysical studies. Here we show that mature fibrils made of 4R tau variants, prepared with heparin or RNA, spontaneously depolymerize and release monomers when their cofactors are removed. We demonstrate that the cross-β-sheet assembly formed in vitro with polyanion addition is unstable at room temperature. We furthermore demonstrate high seeding capacity with transgenic AD mouse brain-extracted tau fibrils in vitro that, however, is exhausted after one generation, while supplementation with RNA cofactors resulted in sustained seeding over multiple generations. We suggest that tau fibrils formed in brains are supported by unknown cofactors and inhere higher-quality packing, as reflected in a more distinct conformational arrangement in the mouse fibril-seeded, compared with heparin-induced, tau fibrils. Our study suggests that the role of cofactors in tauopathies is a worthy focus of future studies, as they may be viable targets for diagnosis and therapeutics.
|
|
|
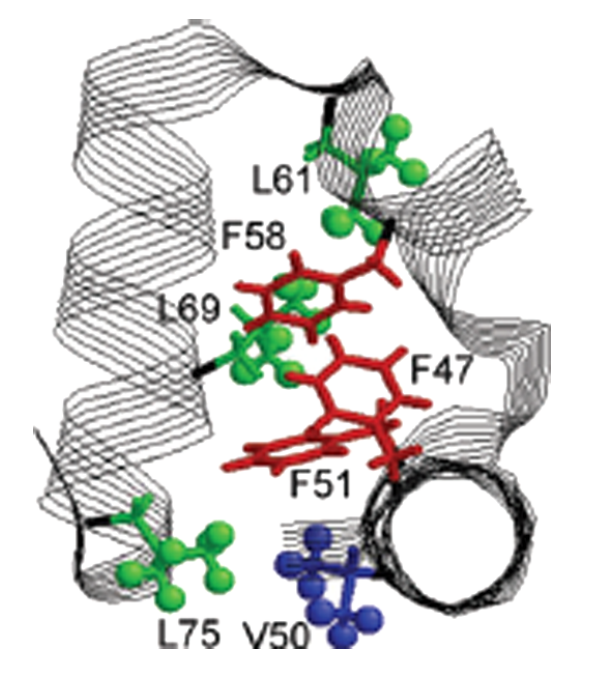
Phenyl-Ring Dynamics in Amyloid Fibrils and Proteins: The Microscopic-Order-Macroscopic-Disorder Perspective
E. Meirovitch, Z. Liang, and J. H. Freed.
J. Phys. Chem. B 122, 8675-8684 (2018)
Supporting Information
<doi: 10.1021/acs.jpcb.8b06330>
PMID:
30141954
PMCID:
PMC6174686
Publication #432
|
|
|
ABSTRACT: We have developed the microscopic-order-macroscopic-disorder (MOMD) approach for studying internal mobility in polycrystalline proteins with 2H lineshape analysis. The motion itself is expressed by a diffusion tensor, R, the local spatial restraints by a potential, u, and the "local geometry" by the relative orientation of the model-related and nuclear magnetic resonance-related tensors. Here, we apply MOMD to phenyl-ring dynamics in several Αβ40-amyloid-fibrils, and the villin headpiece subdomain (HP36). Because the available data are limited in extent and sensitivity, we adjust u and R in the relevant parameter ranges, fixing the "local geometry" in accordance with standard stereochemistry. This yields a physically well-defined and consistent picture of phenyl-ring dynamics, enabling comparison between different systems. In the temperature range of 278–308 K, u has a strength of (1.7–1.8) kT and a rhombicity of (2.4–2.6) kT, and R has components of 5.0 × 102 ≤ R⊥ ≤ 2.0 × 103 s–1 and 6.3 × 105 ≤ R∥ ≤ 2.0 × 106 s–1. At 278 K, fibril hydration increases the axiality of both u and R; HP36 hydration has a similar effect at 295 K, reducing R⊥ considerably. The D23N mutation slows down the motion of the probe; Αβ40 polymorphism affects both this motion and the related local potential. The present study identifies the impact of various factors on phenyl-ring mobility in amyloid fibrils and globular proteins; the difference between the two protein forms is considerable. The distinctive impact of hydration on phenyl-ring motion and previously studied methyl-group motion is also examined. The 2H lineshapes considered here were analyzed previously with various multi-simple-mode (MSM) models, where several simple motional modes are combined. The MOMD and MSM interpretations differ in essence.
|
|
|
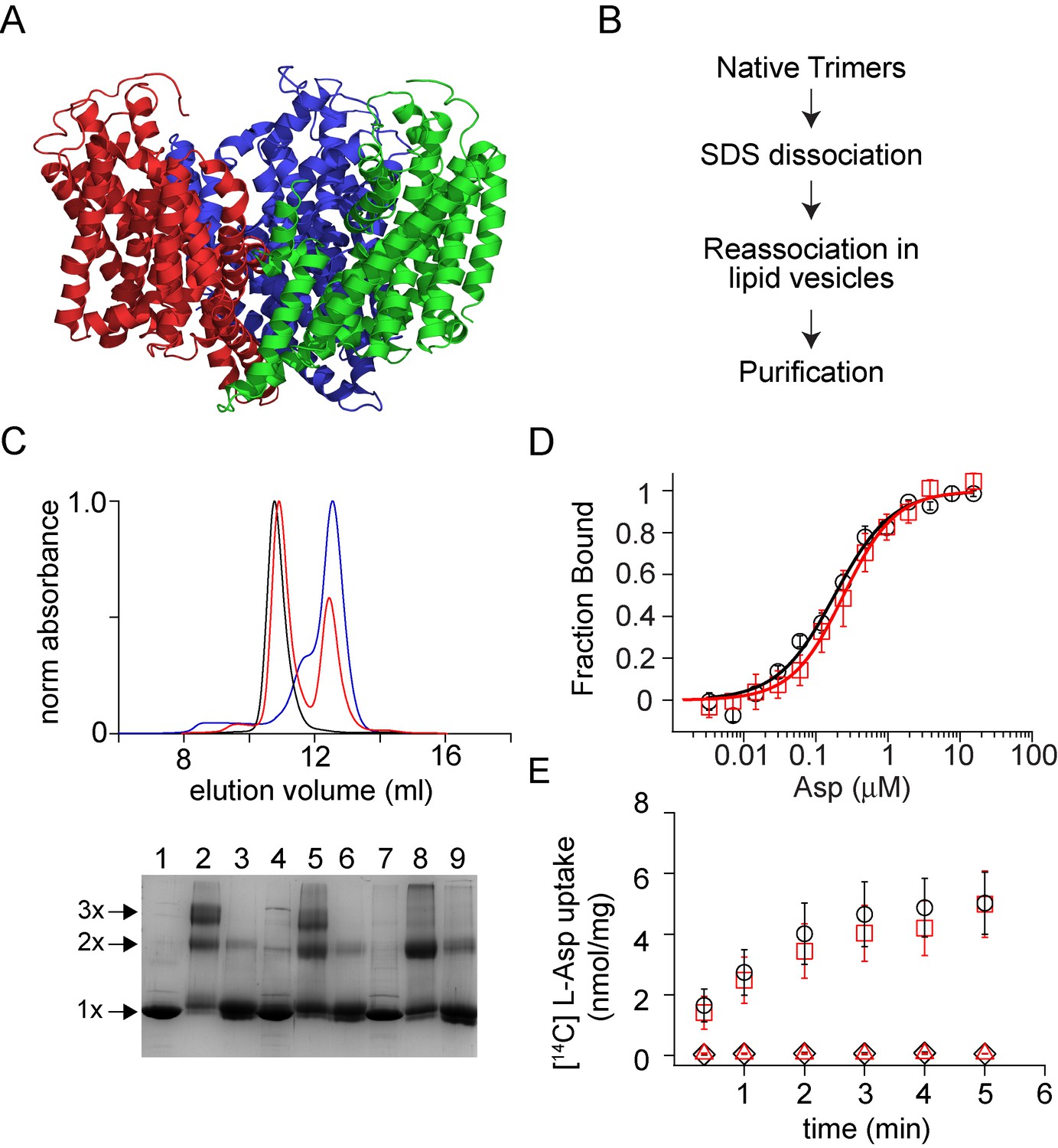
A facile approach for the in vitro assembly of multimeric membrane transport proteins
E. A. Riederer, P. J. Focke, E. R. Georgieva, N. Akyuz, K. Matulef, P. P. Borbat, J. H. Freed, S. C. Blanchard, O. Boudker, and F. I. Valiyaveetil.
eLife 7, e36478 (2018)
|
|
|
A facile approach for the in vitro assembly of multimeric membrane transport proteins
E. A. Riederer, P. J. Focke, E. R. Georgieva, N. Akyuz, K. Matulef, P. P. Borbat, J. H. Freed, S. C. Blanchard, O. Boudker, and F. I. Valiyaveetil.
eLife 7, e36478 (2018)
Supporting Information
<doi: 10.7554/eLife.36478>
PMID:
29889023
PMCID:
PMC6025958
Publication #431
|

|
|
ABSTRACT: Membrane proteins such as ion channels and transporters are frequently homomeric. The homomeric nature raises important questions regarding coupling between subunits and complicates the application of techniques such as FRET or DEER spectroscopy. These challenges can be overcome if the subunits of a homomeric protein can be independently modified for functional or spectroscopic studies. Here, we describe a general approach for in vitro assembly that can be used for the generation of heteromeric variants of homomeric membrane proteins. We establish the approach using GltPh, a glutamate transporter homolog that is trimeric in the native state. We use heteromeric GltPh transporters to directly demonstrate the lack of coupling in substrate binding and demonstrate how heteromeric transporters considerably simplify the application of DEER spectroscopy. Further, we demonstrate the general applicability of this approach by carrying out the in vitro assembly of VcINDY, a Na+-coupled succinate transporter and CLC-ec1, a Cl-/H+ antiporter.
|
|
|
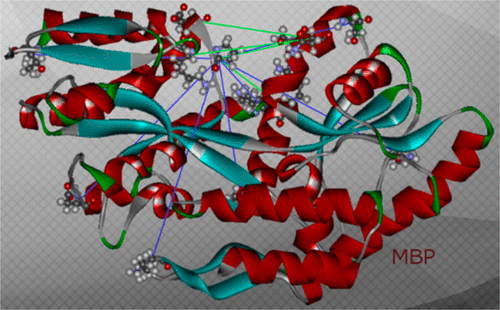
Open and Closed Form of Maltose Binding Protein in Its Native and Molten Globule State As Studied by Electron Paramagnetic Resonance Spectroscopy
B. Selmke, P. P. Borbat, C. Nickolaus, R. Varadarajan, J. H. Freed, and W. E. Trommer.
Biochemistry 57, 5507-5512 (2018)
Supporting Information
<doi: 10.1021/acs.biochem.8b00322>
PMID:
30004675
PMCID:
PMC6211580
Publication #430
|
|
|
ABSTRACT: An intensively investigated intermediate state of protein folding is the molten globule (MG) state, which contains secondary but hardly any tertiary structure. In previous work, we have determined the distances between interacting spins within maltose binding protein (MBP) in its native state using continuous wave and double electron–electron resonance (DEER) electron paramagnetic resonance (EPR) spectroscopy. Seven double mutants had been employed to investigate the structure within the two domains of MBP. DEER data nicely corroborated the previously available X-ray data. Even in its MG state, MBP is known to still bind its ligand maltose. We therefore hypothesized that there must be a defined structure around the binding pocket of MBP already in the absence of tertiary structure. Here we have investigated the functional and structural difference between native and MG state in the open and closed form with a new set of MBP mutants. In these, the spin-label positions were placed near the active site. Binding of its ligands leads to a conformational change from open to closed state, where the two domains are more closely together. The complete set of MBP mutants was analyzed at pH 3.2 (MG) and pH 7.4 (native state) using double-quantum coherence EPR. The values were compared with theoretical predictions of distances between the labels in biradicals constructed by molecular modeling from the crystal structures of MBP in open and closed form and were found to be in excellent agreement. Measurements show a defined structure around the binding pocket of MBP in MG, which explains maltose binding. A new and important finding is that in both states ligand-free MBP can be found in open and closed form, while ligand-bound MBP appears only in closed form because of maltose binding.
|
|
|
Site-Specific Incorporation of a Cu2+ Spin Label into Proteins for Measuring Distances by Pulsed Dipolar Electron Spin Resonance Spectroscopy
G. E. Merz, P. P. Borbat, A. R. Muok, M. Srivastava, D. N. Bunck, J. H. Freed, and B. R. Crane.
J. Phys. Chem. B 122, 9443-9451 (2018)
Supporting Information
<doi: 10.1021/acs.jpcb.8b05619>
PMID:
30222354
PMCID:
PMC6215709
Publication #429
|
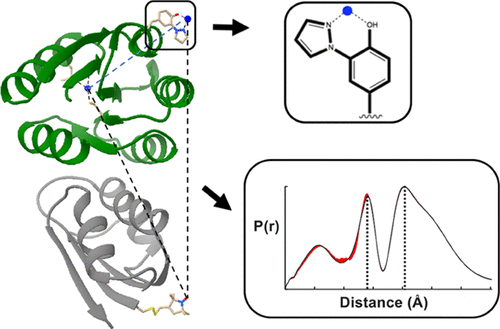
|
|
ABSTRACT: Pulsed dipolar electron spin resonance spectroscopy (PDS) is a powerful tool for measuring distances in solution-state macromolecules. Paramagnetic metal ions, such as Cu2+, are used as spin probes because they can report on metalloprotein features and can be spectroscopically distinguished from traditional nitroxide (NO)-based labels. Here, we demonstrate site-specific incorporation of Cu2+ into non-metalloproteins through the use of a genetically encodable non-natural amino acid, 3-pyrazolyltyrosine (PyTyr). We first incorporate PyTyr in cyan fluorescent protein to measure Cu2+-to-NO distances and examine the effects of solvent conditions on Cu2+ binding and protein aggregation. We then apply the method to characterize the complex formed by the histidine kinase CheA and its target response regulator CheY. The X-ray structure of CheY–PyTyr confirms Cu labeling at PyTyr but also reveals a secondary Cu site. Cu2+-to-NO and Cu2+-to-Cu2+ PDS measurements of CheY–PyTyr with nitroxide-labeled CheA provide new insights into the conformational landscape of the phosphotransfer complex and have implications for kinase regulation.
|
|
|
MOMD Analysis of NMR Line Shapes from Aβ-Amyloid Fibrils: A New Tool for Characterizing Molecular Environments in Protein Aggregates
E. Meirovitch, Z. Liang, and J. H. Freed.
J. Phys. Chem. B 122, 4793-4801 (2018)
Supporting Information
<doi: 10.1021/acs.jpcb.8b02181>
PMID:
29624402
PMCID:
PMC5945340
Publication #428
|
|
|
ABSTRACT: The microscopic-order-macroscopic-disorder (MOMD) approach for 2H NMR line shape analysis is applied to dry and hydrated 3-fold- and 2-fold-symmetric amyloid-Aβ40 fibrils and protofibrils of the D23N mutant. The methyl moieties of L17, L34, V36 (C–CD3), and M35 (S–CD3) serve as probes. Experimental 2H spectra acquired previously in the 147–310 K range are used. MOMD describes local probe motion as axial diffusion (R tensor) in the presence of a potential, u, which represents the spatial restrictions exerted by the molecular surroundings. We find that R∥ = (0.2–3.3) × 104 s–1, R⊥ = (2.2–2.5) × 102 s–1, and R is tilted from the 2H quadrupolar tensor at 60–75°. The strength of u is in the (2.0–2.4) kT range; its rhombicity is substantial. The only methyl moieties affected by fibril hydration are those of M35, located at fibril interfaces. The associated local potentials change form abruptly around 260 K, where massive water freezing occurs. An independent study revealed unfrozen "tightly-peptide-bound" water residing at the interfaces of the 3-fold-symmetric Aβ40 fibrils and at the interfaces of the E22G and E22Δ Aβ40-mutant fibrils. Considering this to be the case in general for Aβ40-related fibrils, the following emerges. The impact of water freezing is transmitted selectively to the fibril structure through interactions with tightly-peptide-bound water, in this case of M35 methyl moieties. The proof that such waters reside at the interfaces of the 2-fold-symmetric fibril, and the protofibril of the D23N mutant, is new. MOMD provides information on the surroundings of the NMR probe directly via the potential, u, which is inherent to the model; a prior interpretation of the same experimental data does so partially and indirectly (see below). Thus, MOMD analysis of NMR line shapes as applied to amyloid fibrils/protein aggregates emerges as a consistent new tool for elucidating the properties of, and processes associated with, molecular environments in the fibril.
|
|
|
Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB
R. S. Cooper, E. R. Georgieva, P. P. Borbat, J. H. Freed, and E. E. Heldwein.
Nat. Struct. Mol. Biol. 25, 416-424 (2018)
Supporting Information
<doi: 10.1038/s41594-018-0060-6>
PMID:
29728654
PMCID:
PMC5942590
Publication #427
|
|
|
ABSTRACT: Viral fusogens merge viral and cell membranes during cell penetration. Their ectodomains drive fusion by undergoing large-scale refolding, but little is known about the functionally important regions located within or near the membrane. Here we report the crystal structure of full-length glycoprotein B (gB), the fusogen from herpes simplex virus, complemented by electron spin resonance measurements. The membrane-proximal (MPR), transmembrane (TMD), and cytoplasmic (CTD) domains form a uniquely folded trimeric pedestal beneath the ectodomain, which balances dynamic flexibility with extensive, stabilizing membrane interactions. The postfusion conformation of the ectodomain suggests that the CTD likewise adopted the postfusion form. However, hyperfusogenic mutations, which destabilize the prefusion state of gB, target key interfaces and structural motifs that reinforce the observed CTD structure. Thus, a similar CTD structure must stabilize gB in its prefusion state. Our data suggest a model for how this dynamic, membrane-dependent 'clamp' controls the fusogenic refolding of gB.
|
|
|
Protein dynamics in the solid-state from 2H NMR lineshape analysis. III. MOMD in the presence of Magic Angle Spinning
E. Meirovitch, Z. Liang, J. H. Freed.
Solid State Nucl. Magn. Reson. 89, 35-44 (2018)
<doi: 10.1016/j.ssnmr.2017.11.001>
PMID:
29208317
PMCID:
PMC5772661
Publication #426
|
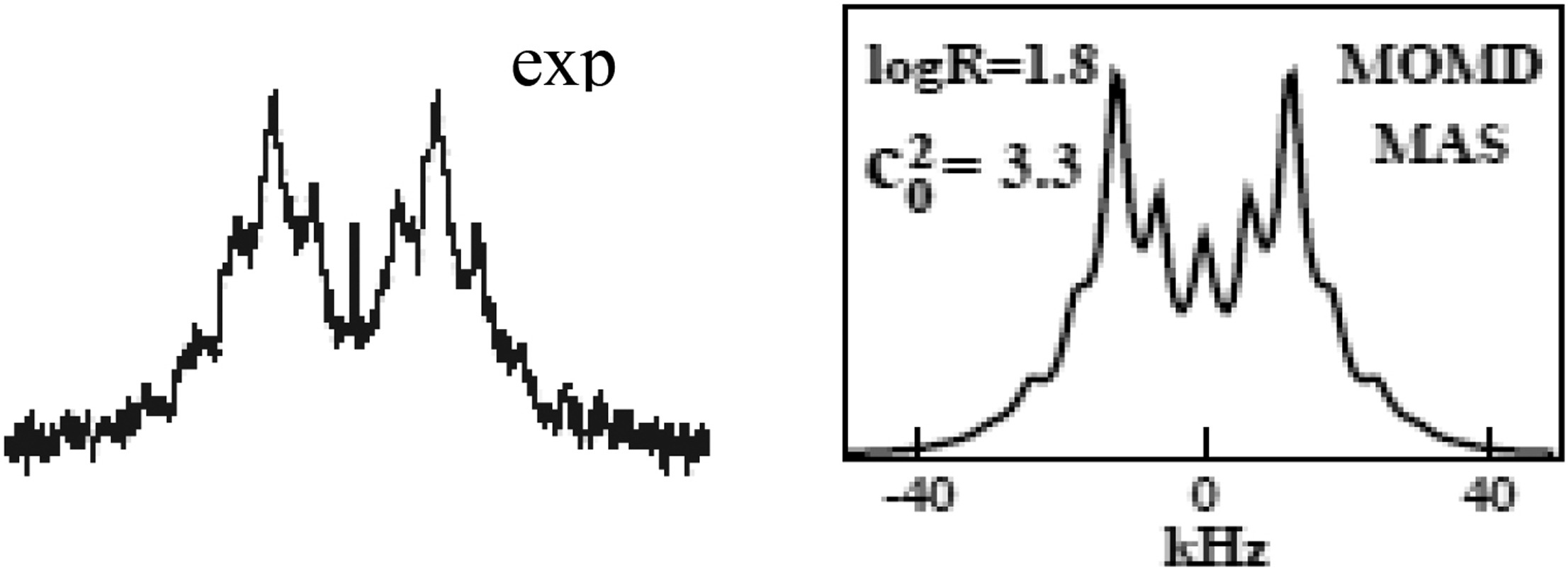
|
|
ABSTRACT: We report on a new approach to the analysis of dynamic NMR lineshapes from polycrystalline (i.e., macroscopically disordered) samples in the presence of Magic Angle Spinning (MAS). This is an application of the Stochastic Liouville Equation developed by Freed and co-workers for treating restricted (i.e., microscopically ordered) motions. The 2H nucleus in an internally-mobile C–CD3 moiety serves as a prototype probe. The acronym is 2H/MOMD/MAS, where MOMD stands for "microscopic-order-macroscopic-disorder." The key elements describing internal motions – their type, the local spatial restrictions, and related features of local geometry – are treated in MOMD generally, within their rigorous three-dimensional tensorial requirements. Based on this representation a single physically well-defined model of local motion has the capability of reproducing experimental spectra. There exist other methods for analyzing dynamic 2H/MAS spectra which advocate simple motional modes. Yet, to reproduce satisfactorily the experimental lineshapes, one has either to use unusual parameter values, or combine several simple motional modes. The multi-simple-mode reasoning assumes independence of the constituent modes, features ambiguity as different simple modes may be used, renders inter-system comparison difficult as the overall models differ, and makes possible model-improvement only by adding yet another simple mode, i.e., changing the overall model. 2H/MOMD/MAS is free of such limitations and inherently provides a clear physical interpretation. These features are illustrated. The advantage of 2H/MOMD/MAS in dealing with sensitive but hardly investigated slow-motional lineshapes is demonstrated by applying it to actual experimental data. The results differ from those obtained previously with a two-site exchange scheme that yielded unusual parameters.
|
|
|
Synthesis and Solution-Phase Characterization of Sulfonated Oligothioetheramides
J. S. Brown, Y. M. Acevedo, G. D. He, J. H. Freed, P. Clancy, and C. A. Alabi.
Macromolecules 50, 8731-8738 (2017)
Supporting Information
<doi: 10.1021/acs.macromol.7b01915>
PMID:
29386690
PMCID:
PMC5788177
Publication #425
|
|
|
ABSTRACT: Nature has long demonstrated the importance of chemical sequence to induce structure and tune physical interactions. Investigating macromolecular structure and dynamics is paramount to understand macromolecular binding and target recognition. To that end, we have synthesized and characterized flexible sulfonated oligothioetheramides (oligoTEAs) by variable temperature pulse field gradient (PFG) NMR, double electron–electron resonance (DEER), and molecular dynamics (MD) simulations to capture their room temperature structure and dynamics in water. We have examined the contributions of synthetic length (2–12mer), pendant group charge, and backbone hydrophobicity. We observe significant entropic collapse, driven in part by backbone hydrophobicity. Analysis of individual monomer contributions revealed larger changes due to the backbone compared to pendant groups. We also observe screening of intramolecular electrostatic repulsions. Finally, we comment on the combination of DEER and PFG NMR measurements via Stokes–Einstein–Sutherland diffusion theory. Overall, this sensitive characterization holds promise to enable de novo development of macromolecular structure and sequence–structure–function relationships with flexible, but biologically functional macromolecules.
|
|
|
Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis
M. Dong, V. Kathiresan, M. K. Fenwick, A. T. Torelli, Y. Zhang, J. D. Caranto, B. Dzikovski, A. Sharma, K. M. Lancaster, J. H. Freed, S. E. Ealick, B. M. Hoffman, and H. Lin.
Science 359, 1247-1250 (2018)
|
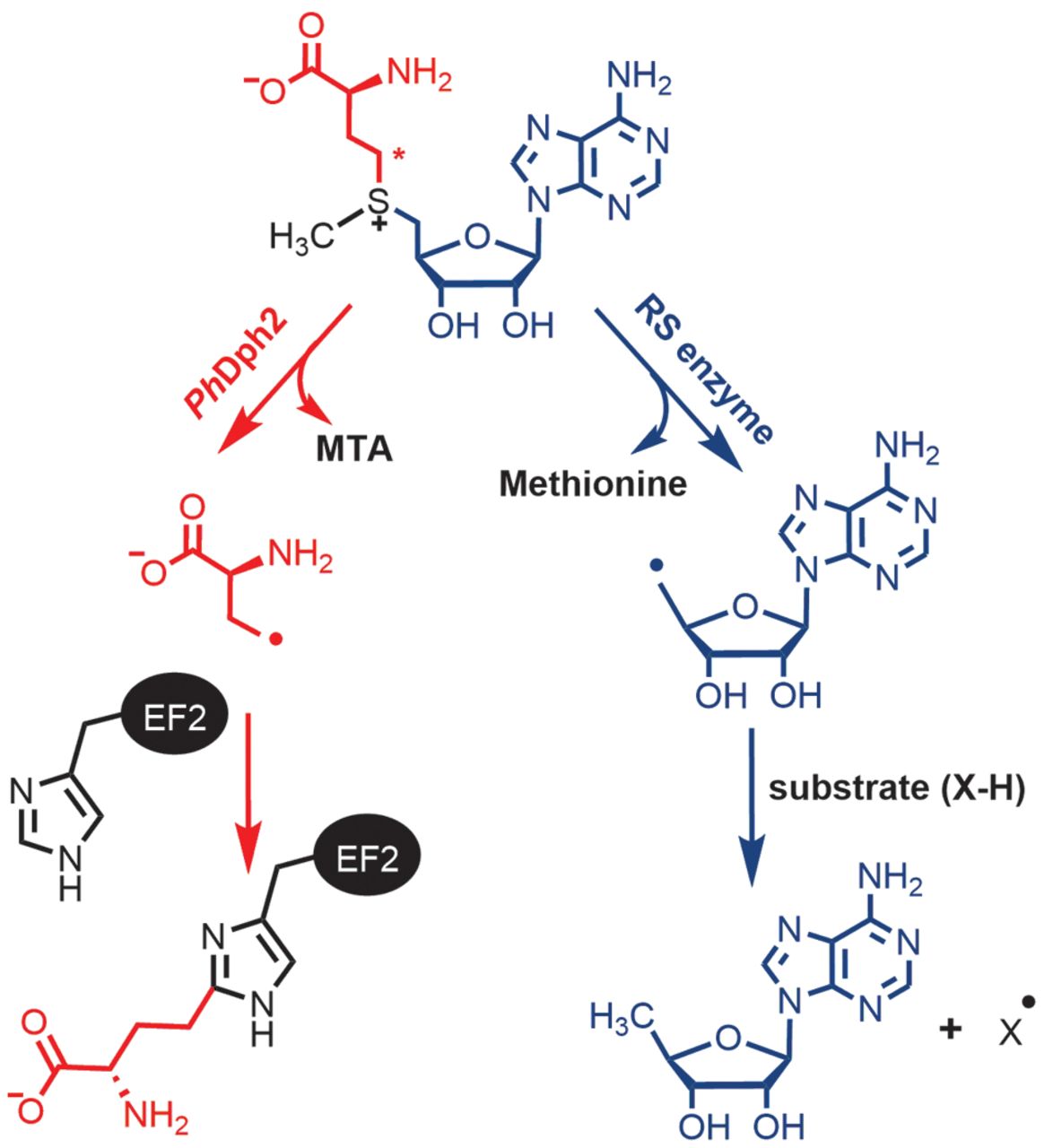
|
|
Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis
M. Dong, V. Kathiresan, M. K. Fenwick, A. T. Torelli, Y. Zhang, J. D. Caranto, B. Dzikovski, A. Sharma, K. M. Lancaster, J. H. Freed, S. E. Ealick, B. M. Hoffman, and H. Lin.
Science 359, 1247-1250 (2018)
Supporting Information
<doi: 10.1126/science.aao6595>
PMID:
29590073
PMCID:
PMC6066404
Publication #424
|

|
|
ABSTRACT: Diphthamide biosynthesis involves a carbon-carbon bond-forming reaction catalyzed by a radical S-adenosylmethionine (SAM) enzyme that cleaves a carbon-sulfur (C–S) bond in SAM to generate a 3-amino-3-carboxypropyl (ACP) radical. Using rapid freezing, we have captured an organometallic intermediate with an iron-carbon (Fe–C) bond between ACP and the enzyme's [4Fe-4S] cluster. In the presence of the substrate protein, elongation factor 2, this intermediate converts to an organic radical, formed by addition of the ACP radical to a histidine side chain. Crystal structures of archaeal diphthamide biosynthetic radical SAM enzymes reveal that the carbon of the SAM C–S bond being cleaved is positioned near the unique cluster Fe, able to react with the cluster. Our results explain how selective C–S bond cleavage is achieved in this radical SAM enzyme.
|
|
|
Dipolar Spectroscopy – Single-Resonance Methods
P. P. Borbat and J. H. Freed.
eMagRes 6 425-462 (2017)
<doi: 10.1002/9780470034590.emrstm1519>
PMID: [None - Book] PMCID:
[None - Book]
Publication #423
|
|
|
ABSTRACT: A range of powerful techniques based on pulse EPR spectroscopies is used extensively for measuring electron spin dipolar couplings between native or introduced paramagnetic reporters, from which accurate distances, and in certain cases molecular orientations, can be determined to aid in solving biomolecular structures. Notably, several versatile pulse EPR methods of high sensitivity were developed over the past two decades. The two main groups of pulse methods are represented by single- and double-resonance techniques, which together constitute pulse dipolar EPR spectroscopy (PDS). We describe the first group by outlining the basic principles and illustrate key techniques with several examples from the literature, as well as some newly produced for purposes of this chapter. Specifically, we describe double-quantum coherence (DQC) methods for robust selection of the dipolar coupling by filtering out undesired spin coherence pathways via phase cycling, and we also discuss single-resonance methods such as four- and five-pulse single-quantum coherence (SQC) methods, all of which benefit from maximizing the spectral coverage. The common features and conceptual differences between single- and double-resonance methods are discussed. In addition, the extensions of modern DQC and SQC methods to two-dimensional correlation spectroscopy for eliciting orientational information are included. Other types of 'single-frequency' PDS methods, not all of which are based on principles behind fully coherent single-resonance methods, are also discussed to show the diversity of PDS methods.
|
|
|
Singular Value Decomposition Method to Determine Distance Distributions in Pulsed Dipolar Electron Spin Resonance
M. Srivastava and J. H. Freed.
J. Phys. Chem. Lett. 8, 5648-5655 (2017)
Supporting Information
<doi: 10.1021/acs.jpclett.7b02379>
PMID:
29099190
PMCID:
PMC5708871
Publication #422
|
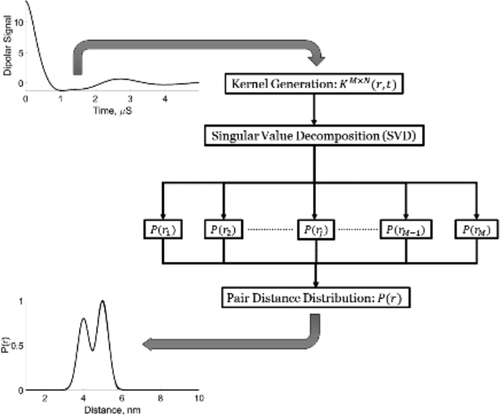
|
|
ABSTRACT: Regularization is often utilized to elicit the desired physical results from experimental data. The recent development of a denoising procedure yielding about 2 orders of magnitude in improvement in SNR obviates the need for regularization, which achieves a compromise between canceling effects of noise and obtaining an estimate of the desired physical results. We show how singular value decomposition (SVD) can be employed directly on the denoised data, using pulse dipolar electron spin resonance experiments as an example. Such experiments are useful in measuring distances and their distributions, P(r) between spin labels on proteins. In noise-free model cases exact results are obtained, but even a small amount of noise (e.g., SNR = 850 after denoising) corrupts the solution. We develop criteria that precisely determine an optimum approximate solution, which can readily be automated. This method is applicable to any signal that is currently processed with regularization of its SVD analysis.
|
|
|
The SARS-CoV Fusion Peptide Forms an Extended Bipartite Fusion Platform that Perturbs Membrane Order in a Calcium-Dependent Manner
A. L. Lai, J. K. Millet, S. Daniel, J. H. Freed, and G. R. Whittaker.
J. Mol. Biol. 429, 3875-3892 (2017)
<doi: 10.1016/j.jmb.2017.10.017>
PMID:
29056462
PMCID:
PMC5705393
Publication #421
|

|
|
ABSTRACT: Coronaviruses (CoVs) are a major infectious disease threat and include the pathogenic human pathogens of zoonotic origin: severe acute respiratory syndrome CoV (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV). Entry of CoVs into host cells is mediated by the viral spike (S) protein, which is structurally categorized as a class I viral fusion protein, within the same group as influenza virus and HIV. However, S proteins have two distinct cleavage sites that can be activated by a much wider range of proteases. The exact location of the CoV fusion peptide (FP) has been disputed. However, most evidence suggests that the domain immediately downstream of the S2′ cleavage site is the FP (amino acids 798–818 SFIEDLLFNKVTLADAGFMKQY for SARS-CoV, FP1). In our previous electron spin resonance spectroscopic studies, the membrane-ordering effect of influenza virus, HIV, and Dengue virus FPs has been consistently observed. In this study, we used this effect as a criterion to identify and characterize the bona fide SARS-CoV FP. Our results indicate that both FP1 and the region immediately downstream (amino acids 816–835 KQYGECLGDINARDLICAQKF, FP2) induce significant membrane ordering. Furthermore, their effects are calcium dependent, which is consistent with in vivo data showing that calcium is required for SARS-CoV S-mediated fusion. Isothermal titration calorimetry showed a direct interaction between calcium cations and both FPs. This Ca2+-dependency membrane ordering was not observed with influenza FP, indicating that the CoV FP exhibits a mechanistically different behavior. Membrane-ordering effects are greater and penetrate deeper into membranes when FP1 and FP2 act in a concerted manner, suggesting that they form an extended fusion "platform."
|
|
|
Mechanistic Insight into the Photocontrolled Cationic Polymerization of Vinyl Ethers
Q. Michaudel, T. Chauviré, V. Kottisch, M. J. Supej, K. J. Stawiasz, L. Shen, W. R. Zipfel, H. D. Abruña, J. H. Freed, and B. P. Fors.
J. Am. Chem. Soc. 139, 15530-15538 (2017)
|
|
|
Mechanistic Insight into the Photocontrolled Cationic Polymerization of Vinyl Ethers
Q. Michaudel, T. Chauviré, V. Kottisch, M. J. Supej, K. J. Stawiasz, L. Shen, W. R. Zipfel, H. D. Abruña, J. H. Freed, and B. P. Fors.
J. Am. Chem. Soc. 139, 15530-15538 (2017)
Supporting Information
<doi: 10.1021/jacs.7b09539>
PMID:
28985061
PMCID:
PMC5806523
Publication #420
|

|
|
ABSTRACT: The mechanism of the recently reported photocontrolled cationic polymerization of vinyl ethers was investigated using a variety of catalysts and chain-transfer agents (CTAs) as well as diverse spectroscopic and electrochemical analytical techniques. Our study revealed a complex activation step characterized by one-electron oxidation of the CTA. This oxidation is followed by mesolytic cleavage of the resulting radical cation species, which leads to the generation of a reactive cation–this species initiates the polymerization of the vinyl ether monomer–and a dithiocarbamate radical that is likely in equilibrium with the corresponding thiuram disulfide dimer. Reversible addition–fragmentation type degenerative chain transfer contributes to the narrow dispersities and control over chain growth observed under these conditions. Finally, the deactivation step is contingent upon the oxidation of the reduced photocatalyst by the dithiocarbamate radical concomitant with the production of a dithiocarbamate anion that caps the polymer chain end. The fine-tuning of the electronic properties and redox potentials of the photocatalyst in both the excited and the ground states is necessary to obtain a photocontrolled system rather than simply a photoinitiated system. The elucidation of the elementary steps of this process will aid the design of new catalytic systems and their real-world applications.
|
|
|
Unique Structural Features of Membrane-Bound C-Terminal Domain Motifs Modulate Complexin Inhibitory Function
D. Snead, A. L. Lai, R. T. Wragg, D. A. Parisotto, T. F. Ramlall, J. S. Dittman, J. H. Freed, and D. Eliezer.
Front. Mol. Neurosci. 10, 154 (2017)
Supporting Information
<doi: 10.3389/fnmol.2017.00154>
PMID:
28596722
PMCID:
PMC5442187
Publication #419
|

|
|
ABSTRACT: Complexin is a small soluble presynaptic protein that interacts with neuronal SNARE proteins in order to regulate synaptic vesicle exocytosis. While the SNARE-binding central helix of complexin is required for both the inhibition of spontaneous fusion and the facilitation of synchronous fusion, the disordered C-terminal domain (CTD) of complexin is specifically required for its inhibitory function. The CTD of worm complexin binds to membranes via two distinct motifs, one of which undergoes a membrane curvature dependent structural transition that is required for efficient inhibition of neurotransmitter release, but the conformations of the membrane-bound motifs remain poorly characterized. Visualizing these conformations is required to clarify the mechanisms by which complexin membrane interactions regulate its function. Here, we employ optical and magnetic resonance spectroscopy to precisely define the boundaries of the two CTD membrane-binding motifs and to characterize their conformations. We show that the curvature dependent amphipathic helical motif features an irregular element of helical structure, likely a pi-bulge, and that this feature is important for complexin inhibitory function in vivo.
|
|
|
Substrate-Dependent Cleavage Site Selection by Unconventional Radical S-Adenosylmethionine Enzymes in Diphthamide Biosynthesis
M. Dong, M. Horitani, B. Dzikovski, J. H. Freed, S. E. Ealick, B. M. Hoffman, and H. Lin.
J. Am. Chem. Soc. 139, 5680-5683 (2017)
Supporting Information
<doi: 10.1021/jacs.7b01712>
PMID:
28383907
PMCID:
PMC5664936
Publication #418
|
|
|
ABSTRACT: S-Adenosylmethionine (SAM) has a sulfonium ion with three distinct C-S bonds. Conventional radical SAM enzymes use a [4Fe-4S] cluster to cleave homolytically the C5′,adenosine-S bond of SAM to generate a 5′-deoxyadenosyl radical, which catalyzes various downstream chemical reactions. Radical SAM enzymes involved in diphthamide biosynthesis, such as Pyrococcus horikoshii Dph2 (PhDph2) and yeast Dph1-Dph2 instead cleave the Cγ,Met-S bond of methionine to generate a 3-amino-3-carboxylpropyl radical. We here show radical SAM enzymes can be tuned to cleave the third C-S bond to the sulfonium sulfur by changing the structure of SAM. With a decarboxyl SAM analogue (dc-SAM), PhDph2 cleaves the Cmethyl-S bond, forming 5′-deoxy-5′-(3-aminopropylthio) adenosine (dAPTA, 1). The methyl cleavage activity, like the cleavage of the other two C-S bonds, is dependent on the presence of a [4Fe-4S]+ cluster. Electron-nuclear double resonance and mass spectroscopy data suggests that mechanistically one of the S atoms in the [4Fe-4S] cluster captures the methyl group from dc-SAM, forming a distinct EPR-active intermediate, which can transfer the methyl group to nucleophiles such as dithiothreitol. This reveals the [4Fe-4S] cluster in a radical SAM enzyme can be tuned to cleave any one of the three bonds to the sulfonium sulfur of SAM or analogues, and is the first demonstration a radical SAM enzyme could switch from an Fe-based one electron transfer reaction to a S-based two electron transfer reaction in a substrate-dependent manner. This study provides an illustration of the versatile reactivity of Fe-S clusters.
|
|
|
Key features of an Hsp70 chaperone allosteric landscape revealed by ion-mobility native mass spectrometry and double electron-electron resonance
A. L. Lai, E. M. Clerico, M. E. Blackburn, N. A. Patel, C. V. Robinson, P. P. Borbat, J. H. Freed, and L. M. Gierasch.
J. Biol. Chem. 292, 8773-8785 (2017)
Supporting Information
<doi: 10.1074/jbc.M116.770404>
PMID:
28428246
PMCID:
PMC5448104
Publication #417
|
|
|
ABSTRACT: Proteins are dynamic entities that populate conformational ensembles, and most functions of proteins depend on their dynamic character. Allostery, in particular, relies on ligand-modulated shifts in these conformational ensembles. Hsp70s are allosteric molecular chaperones with conformational landscapes that involve large rearrangements of their two domains (viz. the nucleotide-binding domain and substrate-binding domain) in response to adenine nucleotides and substrates. However, it remains unclear how the Hsp70 conformational ensemble is populated at each point of the allosteric cycle and how ligands control these populations. We have mapped the conformational species present under different ligand-binding conditions throughout the allosteric cycle of the Escherichia coli Hsp70 DnaK by two complementary methods, ion-mobility mass spectrometry and double electron-electron resonance. Our results obtained under biologically relevant ligand-bound conditions confirm the current picture derived from NMR and crystallographic data of domain docking upon ATP binding and undocking in response to ADP and substrate. Additionally, we find that the helical lid of DnaK is a highly dynamic unit of the structure in all ligand-bound states. Importantly, we demonstrate that DnaK populates a partially docked state in the presence of ATP and substrate and that this state represents an energy minimum on the DnaK allosteric landscape. Because Hsp70s are emerging as potential drug targets for many diseases, fully mapping an allosteric landscape of a molecular chaperone like DnaK will facilitate the development of small molecules that modulate Hsp70 function via allosteric mechanisms.
|
|
|
A New Wavelet Denoising Method for Experimental Time-Domain Signals: Pulsed Dipolar Electron Spin Resonance
M. Srivastava, E. R. Georgieva, and J. H. Freed.
J. Phys. Chem. A 121 2452-2465 (2017)
Supporting Information
<doi: 10.1021/acs.jpca.7b00183>
PMID:
28257206
PMCID:
PMC5553325
Publication #416
|
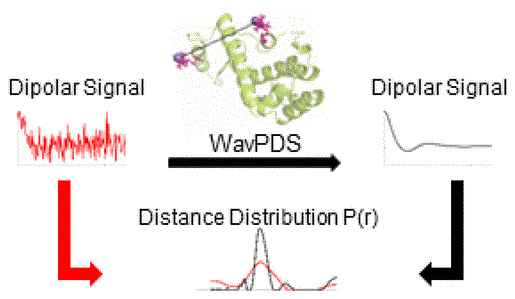
|
|
ABSTRACT: We adapt a new wavelet-transform-based method of denoising experimental signals to pulse-dipolar electron-spin resonance spectroscopy (PDS). We show that signal averaging times of the time-domain signals can be reduced by as much as 2 orders of magnitude, while retaining the fidelity of the underlying signals, in comparison with noiseless reference signals. We have achieved excellent signal recovery when the initial noisy signal has an SNR ≳ 3. This approach is robust and is expected to be applicable to other time-domain spectroscopies. In PDS, these time-domain signals representing the dipolar interaction between two electron spin labels are converted into their distance distribution functions P(r), usually by regularization methods such as Tikhonov regularization. The significant improvements achieved by using denoised signals for this regularization are described. We show that they yield P(r)'s with more accurate detail and yield clearer separations of respective distances, which is especially important when the P(r)'s are complex. Also, longer distance P(r)'s, requiring longer dipolar evolution times, become accessible after denoising. In comparison to standard wavelet denoising approaches, it is clearly shown that the new method (WavPDS) is superior.
|
|
|
Stability and Conformation of a Chemoreceptor HAMP Domain Chimera Correlates with Signaling Properties
N. Sukomon, J. Widom, P. P. Borbat, J. H. Freed, and B. R. Crane.
Biophys. J. 112, 1383-1395 (2017)
Supporting Information
<doi: 10.1016/j.bpj.2017.02.037>
PMID:
28402881
PMCID:
PMC5390053
Publication #415
|
|
|
ABSTRACT: HAMP domains are dimeric, four-helix bundles that transduce conformational signals in bacterial receptors. Genetic studies of the Escherichia coli serine receptor (Tsr) provide an opportunity to understand HAMP conformational behavior in terms of functional output. To increase its stability, the Tsr HAMP domain was spliced into a poly-HAMP unit from the Pseudomonas aeruginosa Aer2 receptor. Within the chimera, the Tsr HAMP undergoes a thermal melting transition at a temperature much lower than that of the Aer2 HAMP domains. Pulse-dipolar electron spin resonance spectroscopy and site-specific spin-labeling confirm that the Tsr HAMP maintains a four-helix bundle. Pulse-dipolar electron spin resonance spectroscopy was also used to study three well-characterized HAMP mutational phenotypes: those that cause flagella rotation that is counterclockwise (CCW) A and kinase-off; CCW B and also kinase-off; and, clockwise (CW) and kinase-on. Conformational properties of the three HAMP variants support a biphasic model of dynamic bundle stability, but also indicate distinct conformational changes within the helix bundle. Functional kinase-on (CW) and kinase-off (CCW A) states differ by concerted changes in the positions of spin-label sites at the base of the bundle. Opposite shifts in the subunit separation distances of neighboring residues at the C-termini of the α1 and α2 helices are consistent with a helix scissors motion or a gearbox rotational model of HAMP activation. In the drastic kinase-off lesion of CCW B, the α1 helices unfold and the α2 helices form a tight two-helix coiled-coil. The substitution of a critical residue in the Tsr N-terminal linker or control cable reduces conformational heterogeneity at the N-terminus of α1 but does not affect structure at the C-terminus of α2. Overall, the data suggest that transitions from on- to off-states involve decreased motional amplitudes of the Tsr HAMP coupled with helix rotations and movements toward a two-helix packing mode.
|
|
|
Structure-Function Studies Link Class II Viral Fusogens with the Ancestral Gamete Fusion Protein HAP2
J. F. Pinello, A. L. Lai, J. K. Millet, D. Cassidy-Hanley, J. H. Freed, and T. G. Clark.
Curr. Biol. 27, 651-660 (2017)
Supporting Information
<doi: 10.1016/j.cub.2017.01.049>
PMID:
28238660
PMCID:
PMC5393271
Publication #414
|
|
|
ABSTRACT: The conserved transmembrane protein, HAP2/GCS1, has been linked to fertility in a wide range of taxa and is hypothesized to be an ancient gamete fusogen. Using template-based structural homology modeling, we now show that the ectodomain of HAP2 orthologs from Tetrahymena thermophila and other species adopt a protein fold remarkably similar to the dengue virus E glycoprotein and related class II viral fusogens. To test the functional significance of this predicted structure, we developed a flow-cytometry-based assay that measures cytosolic exchange across the conjugation junction to rapidly probe the effects of HAP2 mutations in the Tetrahymena system. Using this assay, alterations to a region in and around a predicted "fusion loop" in T. thermophila HAP2 were found to abrogate membrane pore formation in mating cells. Consistent with this, a synthetic peptide corresponding to the HAP2 fusion loop was found to interact directly with model membranes in a variety of biophysical assays. These results raise interesting questions regarding the evolutionary relationships of class II membrane fusogens and harken back to a long-held argument that eukaryotic sex arose as the byproduct of selection for the horizontal transfer of a "selfish" genetic element from cell to cell via membrane fusion.
|
|
|
Signature of an aggregation-prone conformation of tau
N. A. Eschmann, E. R. Georgieva, P. Ganguly, P. P. Borbat, M. D. Rappaport, Y. Akdogan, J. H. Freed, J. -E. Shea, and S. Han.
Sci. Rep. 7, 44739 (2017)
Supporting Information
<doi: 10.1038/srep44739>
PMID:
28303942
PMCID:
PMC5356194
Publication #413
|
|
|
ABSTRACT: The self-assembly of the microtubule associated tau protein into fibrillar cell inclusions is linked to a number of devastating neurodegenerative disorders collectively known as tauopathies. The mechanism by which tau self-assembles into pathological entities is a matter of much debate, largely due to the lack of direct experimental insights into the earliest stages of aggregation. We present pulsed double electron-electron resonance measurements of two key fibril-forming regions of tau, PHF6 and PHF6*, in transient as aggregation happens. By monitoring the end-to-end distance distribution of these segments as a function of aggregation time, we show that the PHF6(*) regions dramatically extend to distances commensurate with extended β-strand structures within the earliest stages of aggregation, well before fibril formation. Combined with simulations, our experiments show that the extended β-strand conformational state of PHF6(*) is readily populated under aggregating conditions, constituting a defining signature of aggregation-prone tau, and as such, a possible target for therapeutic interventions.
|
|
|
Bacterial Energy Sensor Aer Modulates the Activity of the Chemotaxis Kinase CheA Based on the Redox State of the Flavin Cofactor
D. Samanta, J. Widom, P. P. Borbat, J. H. Freed, and B. R. Crane.
J. Biol. Chem. 291, 25809-25814 (2016)
<doi: 10.1074/jbc.C116.757492>
PMID:
27803157
PMCID:
PMC5207056
Publication #412
|
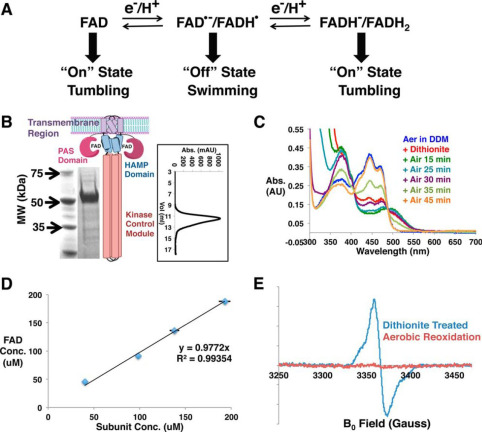
|
|
ABSTRACT: Flagellated bacteria modulate their swimming behavior in response to environmental cues through the CheA/CheY signaling pathway. In addition to responding to external chemicals, bacteria also monitor internal conditions that reflect the availability of oxygen, light, and reducing equivalents, in a process termed "energy taxis." In Escherichia coli, the transmembrane receptor Aer is the primary energy sensor for motility. Genetic and physiological data suggest that Aer monitors the electron transport chain through the redox state of its FAD cofactor. However, direct biochemical data correlating FAD redox chemistry with CheA kinase activity have been lacking. Here, we test this hypothesis via functional reconstitution of Aer into nanodiscs. As purified, Aer contains fully oxidized FAD, which can be chemically reduced to the anionic semiquinone (ASQ). Oxidized Aer activates CheA, whereas ASQ Aer reversibly inhibits CheA. Under these conditions, Aer cannot be further reduced to the hydroquinone, in contrast to the proposed Aer signaling model. Pulse ESR spectroscopy of the ASQ corroborates a potential mechanism for signaling in that the resulting distance between the two flavin-binding PAS (Per-Arnt-Sim) domains implies that they tightly sandwich the signal-transducing HAMP domain in the kinase-off state. Aer appears to follow oligomerization patterns observed for related chemoreceptors, as higher loading of Aer dimers into nanodiscs increases kinase activity. These results provide a new methodological platform to study Aer function along with new mechanistic details into its signal transduction process.
|
|
|
Organometallic Complex Formed by an Unconventional Radical S-Adenosylmethionine Enzyme
M. Dong, M. Horitani, B. Dzikovski, M. -E. Pandelia, C. Krebs, J. H. Freed, B. M. Hoffman, and H. Lin.
J. Am. Chem. Soc. 138, 9755-9758 (2016)
Supporting Information
<doi: 10.1021/jacs.6b04155>
PMID:
27465315
PMCID:
PMC5068486
Publication #411
|

|
|
ABSTRACT: Pyrococcus horikoshii Dph2 (PhDph2) is an unusual radical S-adenosylmethionine (SAM) enzyme involved in the first step of diphthamide biosynthesis. It catalyzes the reaction by cleaving SAM to generate a 3-amino-3-carboxypropyl (ACP) radical. To probe the reaction mechanism, we synthesized a SAM analogue (SAMCA), in which the ACP group of SAM is replaced with a 3-carboxyallyl group. SAMCA is cleaved by PhDph2, yielding a paramagnetic (S = 1/2) species, which is assigned to a complex formed between the reaction product, α-sulfinyl-3-butenoic acid, and the [4Fe-4S] cluster. Electron–nuclear double resonance (ENDOR) measurements with 13C and 2H isotopically labeled SAMCA support a π-complex between the C=C double bond of α-sulfinyl-3-butenoic acid and the unique iron of the [4Fe-4S] cluster. This is the first example of a radical SAM-related [4Fe-4S]+ cluster forming an organometallic complex with an alkene, shedding additional light on the mechanism of PhDph2 and expanding our current notions for the reactivity of [4Fe-4S] clusters in radical SAM enzymes.
|
|
|
A New Wavelet Denoising Method for Selecting Decomposition Levels and Noise Thresholds
M. Srivastava, C. L. Anderson, and J. H. Freed.
IEEE Access 4 3862-3877 (2016)
<doi: 10.1109/ACCESS.2016.2587581>
PMID:
27795877
PMCID:
PMC5079185
Publication #410
|
|
|
ABSTRACT: A new method is presented to denoise 1-D experimental signals using wavelet transforms. Although the state-of-the-art wavelet denoising methods perform better than other denoising methods, they are not very effective for experimental signals. Unlike images and other signals, experimental signals in chemical and biophysical applications, for example, are less tolerant to signal distortion and under-denoising caused by the standard wavelet denoising methods. The new method: 1) provides a method to select the number of decomposition levels to denoise; 2) uses a new formula to calculate noise thresholds that does not require noise estimation; 3) uses separate noise thresholds for positive and negative wavelet coefficients; 4) applies denoising to the approximation component; and 5) allows the flexibility to adjust the noise thresholds. The new method is applied to continuous wave electron spin resonance spectra and it is found that it increases the signal-to-noise ratio (SNR) by more than 32 dB without distorting the signal, whereas standard denoising methods improve the SNR by less than 10 dB and with some distortion. In addition, its computation time is more than six times faster.
|
|
|
Conformational Response of Influenza A M2 Transmembrane Domain to Amantadine Drug Binding at Low pH (pH 5.5)
E. R. Georgieva, P. P. Borbat, K. Grushin, S. Stoilova-McPhie, N. J. Kulkarni, Z. Liang, and J. H. Freed.
Front. Physiol. 7, 317 (2016)
Supporting Information
<doi: 10.3389/fphys.2016.00317>
PMID:
27524969
PMCID:
PMC4965473
Publication #409
|
|
|
ABSTRACT: The M2 protein from influenza A plays important roles in its viral cycle. It contains a single transmembrane helix, which oligomerizes into a homotetrameric proton channel that conducts in the low-pH environment of the host-cell endosome and Golgi apparatus, leading to virion uncoating at an early stage of infection. We studied conformational rearrangements that occur in the M2 core transmembrane domain residing on the lipid bilayer, flanked by juxtamembrane residues (M2TMD21–49 fragment), upon its interaction with amantadine drug at pH 5.5 when M2 is conductive. We also tested the role of specific mutation and lipid chain length. Electron spin resonance (ESR) spectroscopy and electron microscopy were applied to M2TMD21–49, labeled at the residue L46C with either nitroxide spin-label or Nanogold® reagent, respectively. Electron microscopy confirmed that M2TMD21–49 reconstituted into DOPC/POPS at 1:10,000 peptide-to-lipid molar ratio (P/L) either with or without amantadine, is an admixture of monomers, dimers, and tetramers, confirming our model based on a dimer intermediate in the assembly of M2TMD21–49. As reported by double electron-electron resonance (DEER), in DOPC/POPS membranes amantadine shifts oligomer equilibrium to favor tetramers, as evidenced by an increase in DEER modulation depth for P/L's ranging from 1:18,000 to 1:160. Furthermore, amantadine binding shortens the inter-spin distances (for nitroxide labels) by 5–8 Å, indicating drug induced channel closure on the C-terminal side. No such effect was observed for the thinner membrane of DLPC/DLPS, emphasizing the role of bilayer thickness. The analysis of continuous wave (cw) ESR spectra of spin-labeled L46C residue provides additional support to a more compact helix bundle in amantadine-bound M2TMD21–49 through increased motional ordering. In contrast to wild-type M2TMD21–49, the amantadine-bound form does not exhibit noticeable conformational changes in the case of G34A mutation found in certain drug-resistant influenza strains. Thus, the inhibited M2TMD21–49 channel is a stable tetramer with a closed C-terminal exit pore. This work is aimed at contributing to the development of structure-based anti-influenza pharmaceuticals.
|
|
|
Local Ordering at Mobile Sites in Proteins from Nuclear Magnetic Resonance Relaxation: the Role of Site Symmetry
O. Tchaicheeyan, J. H. Freed, and E. Meirovitch.
J. Phys. Chem. B 120 2886-2898 (2016)
Supporting Information
<doi: 10.1021/acs.jpcb.6b00524>
PMID:
26938937
PMCID:
PMC4837759
Publication #408
|
|
|
ABSTRACT: Restricted motions in proteins (e.g., N–H bond dynamics) are studied effectively with NMR. By analogy with restricted motions in liquid crystals (LC), the local ordering has in the past been primarily represented by potentials comprising the L = 2, |K| = 0, 2 spherical harmonics. However, probes dissolved in LCs experience nonpolar ordering, often referred to as alignment, while protein-anchored probes experience polar ordering, often referred to as orientation. In this study we investigate the role of local (site) symmetry in the context of the polarity of the local ordering. We find that potentials comprising the L = 1, |K| = 0, 1 spherical harmonics represent adequately polar ordering. It is useful to characterize potential symmetry in terms of the irreducible representations of D2h point group, which is already implicit in the definition of the rotational diffusion tensor. Thus, the relevant rhombic L = 1 potentials have B1u and B3u symmetry whereas the relevant rhombic L = 2 potentials have Ag symmetry. A comprehensive scheme where local potentials and corresponding probability density functions (PDFs) are represented in Cartesian and spherical coordinates clarifies how they are affected by polar and nonpolar ordering. The Cartesian coordinates are chosen so that the principal axis of polar axial PDF is pointing along the z-axis, whereas the principal axis of the nonpolar axial PDF is pointing along ±z. Two-term axial potentials with 1 ≤ L ≤ 3 exhibit substantial diversity; they are expected to be useful in NMR-relaxation-data-fitting. It is shown how potential coefficients are reflected in the experimental order parameters. The comprehensive scheme representing local potentials and PDFs is exemplified for the L = 2 case using experimental data from 15N-labeled plexin-B1 and thioredoxin, 2H-, and 13C-labeled benzenehexa-n-alkanoates, and nitroxide-labeled T4 lysozyme. Future prospects for improved ordering analysis based on combined atomistic and mesoscopic approaches are delineated.
|
|
|
Structural Basis for Activation, Assembly and Membrane Binding of ESCRT-III Snf7 Filaments
S. Tang, W. M. Henne, P. P. Borbat, N. J. Buchkovich, J. H. Freed, Y. Mao, J. C. Fromme, and S. D. Emr.
eLife 4 e12548 (2015)
Supporting Information
<doi: 10.7554/eLife.12548>
PMID:
26670543
PMCID:
PMC4720517
Publication #407
|
|
|
ABSTRACT: The endosomal sorting complexes required for transport (ESCRTs) constitute hetero-oligomeric machines that catalyze multiple topologically similar membrane-remodeling processes. Although ESCRT-III subunits polymerize into spirals, how individual ESCRT-III subunits are activated and assembled together into a membrane-deforming filament remains unknown. Here, we determine X-ray crystal structures of the most abundant ESCRT-III subunit Snf7 in its active conformation. Using pulsed dipolar electron spin resonance spectroscopy (PDS), we show that Snf7 activation requires a prominent conformational rearrangement to expose protein-membrane and protein-protein interfaces. This promotes the assembly of Snf7 arrays with ˜30 Å periodicity into a membrane-sculpting filament. Using a combination of biochemical and genetic approaches, both in vitro and in vivo, we demonstrate that mutations on these protein interfaces halt Snf7 assembly and block ESCRT function. The architecture of the activated and membrane-bound Snf7 polymer provides crucial insights into the spatially unique ESCRT-III-mediated membrane remodeling.
|
|
|
Pulsed Dipolar Spectroscopy Reveals that Tyrosyl Radicals Are Generated in Both Monomers of the Cyclooxygenase-2 Dimer
B. J. Orlando, P. P. Borbat, E. R. Georgieva, J. H. Freed, and M. G. Malkowski.
Biochemistry 54 7309-7312 (2015)
Supporting Information
<doi: 10.1021/acs.biochem.5b00979>
PMID:
26636181
PMCID:
PMC4707933
Publication #406
|
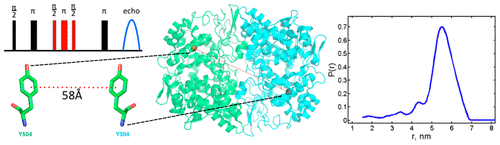
|
|
ABSTRACT: Cyclooxygenases (COXs) are heme-containing sequence homodimers that utilize tyrosyl radical-based catalysis to oxygenate substrates. Tyrosyl radicals are formed from a single turnover of substrate in the peroxidase active site generating an oxy-ferryl porphyrin cation radical intermediate that subsequently gives rise to a Tyr-385 radical in the cyclooxygenase active site and a Tyr-504 radical nearby. We have utilized double-quantum coherence (DQC) spectroscopy to determine the distance distributions between Tyr-385 and Tyr-504 radicals in COX-2. The distances obtained with DQC confirm that Tyr-385 and Tyr-504 radicals were generated in each monomer and accurately match the distances measured in COX-2 crystal structures.
|
|
|
Protein Dynamics in the Solid State from 2H NMR Line Shape Analysis. II. MOMD Applied to C–D and C–CD3 Probes
E. Meirovitch, Z. Liang, and J. H. Freed.
J. Phys. Chem. B 119 14022-14032 (2015)
<doi: 10.1021/acs.jpcb.5b07434>
PMID:
26402431
PMCID:
PMC4676681
Publication #405
|

|
|
ABSTRACT: Deuterium line shape analysis from mobile C–D and C–CD3 groups has emerged as a particularly useful tool for studying dynamics in the solid state. The theoretical models devised so far consist typically of sets of independent dynamic modes. Each such mode is simple and usually case-specific. In this scenario, model improvement entails adding yet another mode (thereby changing the overall model), comparison of different cases is difficult, and ambiguity is unavoidable. We recently developed the microscopic order macroscopic disorder (MOMD) approach as a single-mode alternative. In MOMD, the local spatial restrictions are expressed by an anisotropic potential, the local motion by a diffusion tensor, and the local molecular geometry by relative (magnetic and model-related) tensor orientations, all of adjustable symmetry. This approach provides a consistent method of analysis, thus resolving the issues above. In this study, we apply MOMD to PS-adsorbed LKα14 peptide and dimethylammonium tetraphenylborate (C–CD3 and N–CD3 dynamics, respectively), as well as HhaI methyltransferase target DNA and phase III of benzene-6-hexanoate (C–D dynamics). The success with fitting these four disparate cases, as well as the two cases in the previous report, demonstrates the generality of this MOMD-based approach. In this study, C–D and C–CD3 are both found to execute axial diffusion (rates R⊥ and R∥) in the presence of a rhombic potential given by the L = 2 spherical harmonics (coefficients c02 and c22). R⊥ (R∥) is in the 102–103 (104–105) s–1 range, and c02 and c22 are on the order of 2–3 kBT. Specific parameter values are determined for each mobile site. The diffusion and quadrupolar tensors are tilted at either 120° (consistent with trans–gauche isomerization) or nearly 110.5° (consistent with methyl exchange). Future prospects include extension of the MOMD formalism to include MAS, and application to 15N and 13C nuclei.
|
|
|
Study of paramagnetic defect centers in as-grown and annealed TiO2 anatase and rutile nanoparticles by a variable-temperature X-band and high-frequency (236 GHz) EPR
S. K. Misra, S. I. Andronenko, D. Tipikin, J. H. Freed, V. Somani, and O. Prakash.
J. Magn. Magn. Mater. 401, 495-505 (2016)
<doi: 10.1016/j.jmmm.2015.10.072>
PMID:
27041794
PMCID:
PMC4815036
Publication #404
|

|
|
ABSTRACT: Detailed EPR investigations on as-grown and annealed TiO2 nanoparticles in the anatase and rutile phases were carried out at X-band (9.6 GHz) at 77, 120–300 K and at 236 GHz at 292 K. The analysis of EPR data for as-grown and annealed anatase and rutile samples revealed the presence of several paramagnetic centers: Ti3+, O−, adsorbed oxygen (O2−) and oxygen vacancies. On the other hand, in as-grown rutile samples, there were observed EPR lines due to adsorbed oxygen (O2−) and the Fe3+ ions in both Ti4+ substitutional positions, with and without coupling to an oxygen vacancy in the near neighborhood. Anatase nanoparticles were completely converted to rutile phase when annealed at 1000° C, exhibiting EPR spectra similar to those exhibited by the as-grown rutile nanoparticles. The high-frequency (236 GHz) EPR data on anatase and rutile samples, recorded in the region about g=2.0 exhibit resolved EPR lines, due to O− and O2− ions enabling determination of their g-values with higher precision, as well as observation of hyperfine sextets due to Mn2+ and Mn4+ ions in anatase.
|
|
|
Interaction of Spin-Labeled Lipid Membranes with Transition Metal Ions
B. Dzikovski, V. Livshits, and J.H. Freed
J. Phys. Chem. B 119, 13330-13346 (2015)
Supporting Information
<doi: 10.1021/acs.jpcb.5b08165>
PMID:
26490692
PMCID:
PMC4762260
Publication #403
|
|
|
ABSTRACT: The large values of spin relaxation enhancement (RE) for PC spin-labels in the phospholipid membrane induced by paramagnetic metal salts dissolved in the aqueous phase can be explained by Heisenberg spin exchange due to conformational fluctuations of the nitroxide group as a result of membrane fluidity, flexibility of lipid chains, and, possibly, amphiphilic nature of the nitroxide label. Whether the magnetic interaction occurs predominantly via Heisenberg spin exchange (Ni) or by the dipole–dipole (Gd) mechanism, it is essential for the paramagnetic ion to get into close proximity to the nitroxide moiety for efficient RE. For different salts of Ni the RE in phosphatidylcholine membranes follows the anionic Hofmeister series and reflects anion adsorption followed by anion-driven attraction of paramagnetic cations on the choline groups. This adsorption is higher for chaotropic ions, e.g., perchlorate. (A chaotropic agent is a molecule in water solution that can disrupt the hydrogen bonding network between water molecules.) However, there is no anionic dependence of RE for model membranes made from negatively charged lipids devoid of choline groups. We used Ni-induced RE to study the thermodynamics and electrostatics of ion/membrane interactions. We also studied the effect of membrane composition and the phase state on the RE values. In membranes with cholesterol a significant difference is observed between PC labels with nitroxide tethers long enough vs not long enough to reach deep into the membrane hydrophobic core behind the area of fused cholesterol rings. This study indicates one must be cautious in interpreting data obtained by PC labels in fluid membranes in terms of probing membrane properties at different immersion depths when it can be affected by paramagnetic species at the membrane surface.
|
|
|
Signal Transduction in Light–Oxygen–Voltage Receptors Lacking the Adduct-Forming Cysteine Residue
E.F. Yee, R.P. Diensthuber, A.T. Vaidya, P.P. Borbat, C. Engelhard, J.H. Freed, R. Bittl, A. Möglich, and B.R. Crane.
Nature Comm. 6 10079 (2015)
Supporting Information
<doi: 10.1038/ncomms10079>
PMID:
26648256
PMCID:
PMC4682037
Publication #402
|

|
|
ABSTRACT: Light–oxygen–voltage (LOV) receptors sense blue light through the photochemical generation of a covalent adduct between a flavin-nucleotide chromophore and a strictly conserved cysteine residue. Here we show that, after cysteine removal, the circadian-clock LOV-protein Vivid still undergoes light-induced dimerization and signalling because of flavin photoreduction to the neutral semiquinone (NSQ). Similarly, photoreduction of the engineered LOV histidine kinase YF1 to the NSQ modulates activity and downstream effects on gene expression. Signal transduction in both proteins hence hinges on flavin protonation, which is common to both the cysteinyl adduct and the NSQ. This general mechanism is also conserved by natural cysteine-less, LOV-like regulators that respond to chemical or photoreduction of their flavin cofactors. As LOV proteins can react to light even when devoid of the adduct-forming cysteine, modern LOV photoreceptors may have arisen from ancestral redox-active flavoproteins. The ability to tune LOV reactivity through photoreduction may have important implications for LOV mechanism and optogenetic applications.
|
|
|
The Interaction between Influenza HA Fusion Peptide and Transmembrane Domain Affects Membrane Structure
A.L. Lai and J.H. Freed
Biophys. J. 109, 2523-2536 (2015)
Supporting Information
<doi: 10.1016/j.bpj.2015.10.044>
PMID:
26682811
PMCID:
PMC4699882
Publication #401
|

|
|
ABSTRACT: Viral glycoproteins, such as influenza hemagglutinin (HA) and human immunodeficiency virus gp41, are anchored by a single helical segment transmembrane domain (TMD) on the viral envelope membrane. The fusion peptides (FP) of the glycoproteins insert into the host membrane and initiate membrane fusion. Our previous study showed that the FP or TMD alone perturbs membrane structure. Interaction between the influenza HA FP and TMD has previously been shown, but its role is unclear. We used PC spin labels dipalmitoylphospatidyl-tempo-choline (on the headgroup), 5PC and 14PC (5-C and 14-C positions on the acyl chain) to detect the combined effect of FP-TMD interaction by titrating HA FP to TMD-reconstituted 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dimyristoyl-sn-glycero-3-phospho-(1'-rac-glycerol)/cholesterol lipid bilayers using electron spin resonance. We found that the FP-TMD increases the lipid order at all positions, which has a greater lipid ordering effect than the sum of the FP or TMD alone, and this effect reaches deeper into the membranes. Although HA-mediated membrane fusion is pH dependent, this combined effect is observed at both pH 5 and pH 7. In addition to increasing lipid order, multiple components are found for 5PC at increased concentration of FP-TMD, indicating that distinct domains are induced. However, the mutation of Gly1 in the FP and L187 in the TMD eliminates the perturbations, consistent with their fusogenic phenotypes. Electron spin resonance on spin-labeled peptides confirms these observations. We suggest that this interaction may provide a driving force in different stages of membrane fusion: initialization, transition from hemifusion stalk to transmembrane contact, and fusion pore formation.
|
|
|
Preformed Soluble Chemoreceptor Trimers That Mimic Cellular Assembly States and Activate CheA Autophosphorylation
A.R. Greenswag, X. Li, P.P. Borbat, D. Samanta, K.J. Watts, J.H. Freed, and B.R. Crane
Biochemistry 54, 3454-3468 (2015).
Supporting Information
<doi: 10.1021/bi501570n>
PMID:
25967982
PMCID:
PMC4772074
Publication #400
|
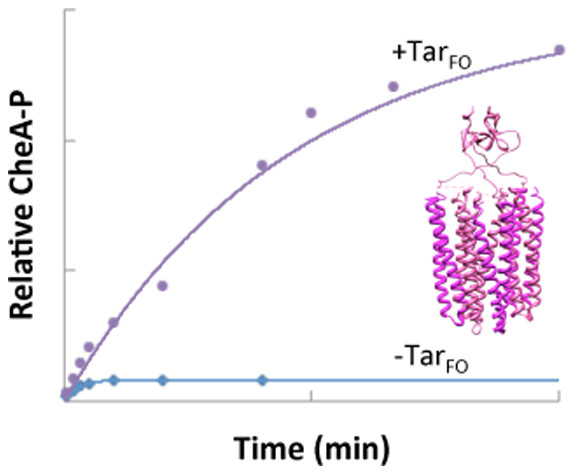
|
|
ABSTRACT: Bacterial chemoreceptors associate with the histidine kinase CheA and coupling protein CheW to form extended membrane arrays that receive and transduce environmental signals. A receptor trimers-of-dimers resides at each vertex of the hexagonal protein lattice. CheA is fully activated and regulated when it is integrated into the receptor assembly. To mimic these states in solution, we have engineered chemoreceptor cytoplasmic kinase-control modules (KCMs) based on the Escherichia coli aspartate receptor Tar that are covalently fused and trimerized by a foldon domain (TarFO). Small-angle X-ray scattering, multi-angle light scattering, and pulsed-dipolar electron spin resonance spectroscopy of spin-labeled proteins indicate that the TarFO modules assemble into homogeneous trimers wherein the protein interaction regions closely associate at the end opposite to the foldon domains. The TarFO variants greatly increase the saturation levels of phosphorylated CheA (CheA-P), indicating that the association with a trimer of receptor dimers changes the fraction of active kinase. However, the rate constants for CheA-P formation with the Tar variants are low compared to those for autophosphorylation by free CheA, and net phosphotransfer from CheA to CheY does not increase commensurately with CheA autophosphorylation. Thus, the Tar variants facilitate slow conversion to an active form of CheA that then undergoes stable autophosphorylation and is capable of subsequent phosphotransfer to CheY. Free CheA is largely incapable of phosphorylation but contains a small active fraction. Addition of TarFO to CheA promotes a planar conformation of the regulatory domains consistent with array models for the assembly state of the ternary complex and different from that observed with a single inhibitory receptor. Introduction of TarFO into E. coli cells activates endogenous CheA to produce increased clockwise flagellar rotation, with the effects increasing in the presence of the chemotaxis methylation system (CheB/CheR). Overall, the TarFO modules demonstrate that trimerized signaling tips self-associate, bind CheA and CheW, and facilitate conversion of CheA to an active conformation.
|
|
|
Focus: Two-dimensional electron-electron double resonance and molecular motions: The challenge of higher frequencies
J.M. Franck, S. Chandrasekaran, B. Dzikovski, C.R. Dunnam, and J.H. Freed.
J. Chem. Phys. 142, 212302-212314 (2015).
<doi: 10.1063/1.4917322>
PMID:
26049420
PMCID:
PMC4443839
Publication #399
|
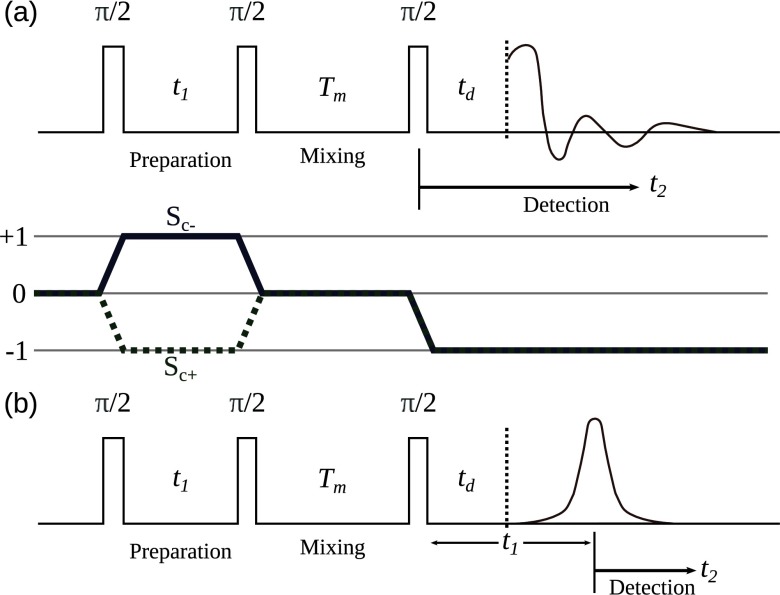
|
|
ABSTRACT: The development, applications, and current challenges of the pulsed ESR technique of two-dimensional Electron-Electron Double Resonance (2D ELDOR) are described. This is a three-pulse technique akin to 2D Exchange Nuclear Magnetic Resonance, but involving electron spins, usually in the form of spin-probes or spin-labels. As a result, it required the extension to much higher frequencies, i.e., microwaves, and much faster time scales, with π/2 pulses in the 2-3 ns range. It has proven very useful for studying molecular dynamics in complex fluids, and spectral results can be explained by fitting theoretical models (also described) that provide a detailed analysis of the molecular dynamics and structure. We discuss concepts that also appear in other forms of 2D spectroscopy but emphasize the unique advantages and difficulties that are intrinsic to ESR. Advantages include the ability to tune the resonance frequency, in order to probe different motional ranges, while challenges include the high ratio of the detection dead time vs. the relaxation times. We review several important 2D ELDOR studies of molecular dynamics. (1) The results from a spin probe dissolved in a liquid crystal are followed throughout the isotropic → nematic → liquid-like smectic → solid-like smectic → crystalline phases as the temperature is reduced and are interpreted in terms of the slowly relaxing local structure model. Here, the labeled molecule is undergoing overall motion in the macroscopically aligned sample, as well as responding to local site fluctuations. (2) Several examples involving model phospholipid membranes are provided, including the dynamic structural characterization of the boundary lipid that coats a transmembrane peptide dimer. Additionally, subtle differences can be elicited for the phospholipid membrane phases: liquid disordered, liquid ordered, and gel, and the subtle effects upon the membrane, of antigen cross-linking of receptors on the surface of plasma membrane, vesicles can be observed. These 2D ELDOR experiments are performed as a function of mixing time, Tm, i.e., the time between the second and third π/2 pulses, which provides a third dimension. In fact, a fourth dimension may be added by varying the ESR frequency/magnetic field combination. Therefore, (3) it is shown how continuous-wave multifrequency ESR studies enable the decomposition of complex dynamics of, e.g., proteins by virtue of their respective time scales. These studies motivate our current efforts that are directed to extend 2D ELDOR to higher frequencies, 95 GHz in particular (from 9 and 17 GHz), in order to enable multi-frequency 2D ELDOR. This required the development of quasi-optical methods for performing the mm-wave experiments, which are summarized. We demonstrate state-of-the-art 95 GHz 2D ELDOR spectroscopy through its ability to resolve the two signals from a spin probe dissolved in both the lipid phase and the coexisting aqueous phase. As current 95 GHz experiments are restricted by limited spectral coverage of the π/2 pulse, as well as the very short T2 relaxation times of the electron spins, we discuss how these limitations are being addressed.
|
|
|
Mechanism of Influenza A M2 Transmembrane Domain Assembly in Lipid Membranes
E.R. Georgieva, P.P. Borbat, H.D. Norman, and J.H. Freed
Sci. Rep. 5, 11757 (2015).
Supporting Information
<doi: 10.1038/srep11757>
PMID:
26190831
PMCID:
PMC4507135
Publication #398
|
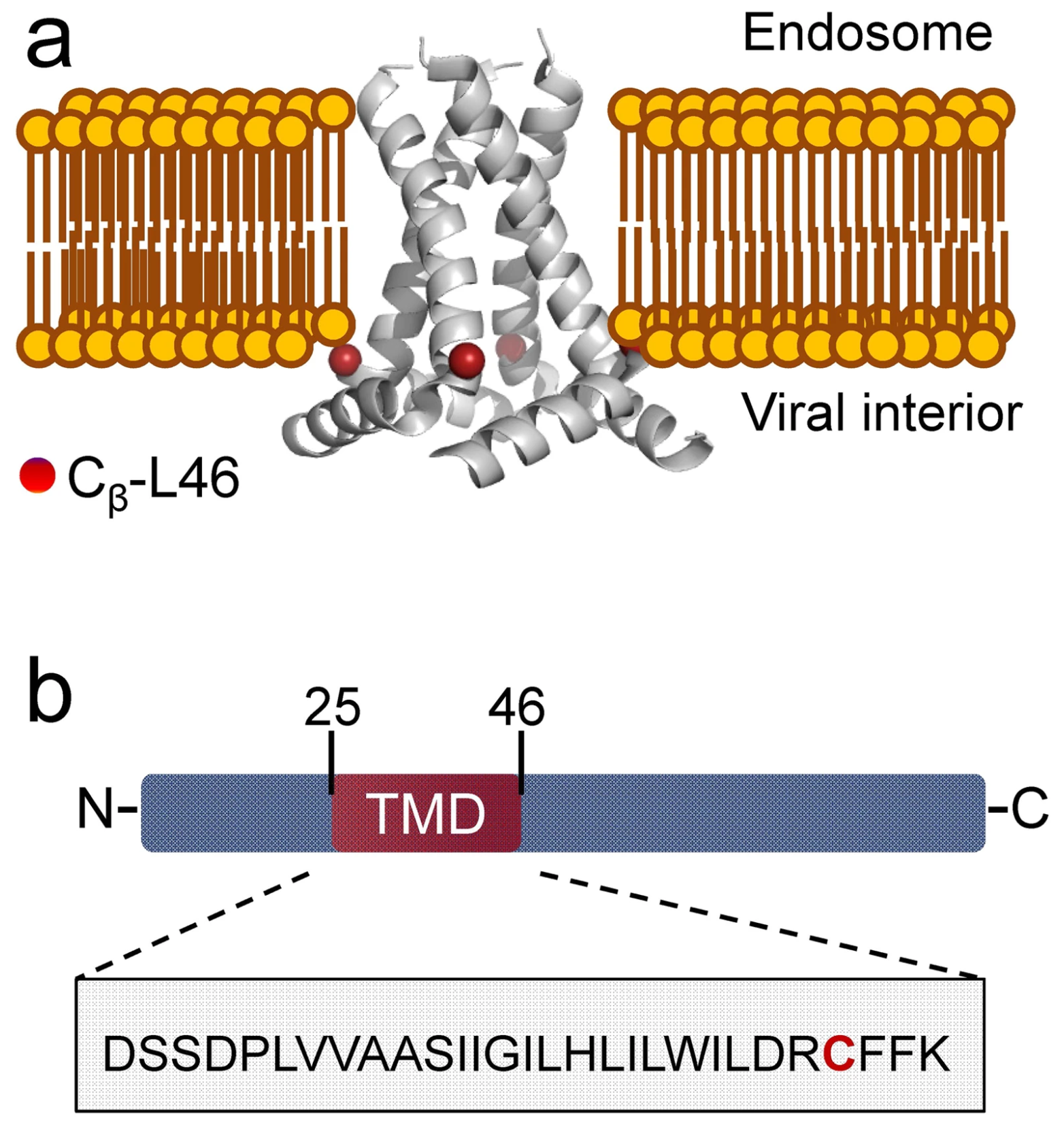
|
|
ABSTRACT: M2 from influenza A virus functions as an oligomeric proton channel essential for the viral cycle, hence it is a high-priority pharmacological target whose structure and functions require better understanding. We studied the mechanism of M2 transmembrane domain (M2TMD) assembly in lipid membranes by the powerful biophysical technique of double electron-electron resonance (DEER) spectroscopy. By varying the M2TMD-to-lipid molar ratio over a wide range from 1:18,800 to 1:160, we found that M2TMD exists as monomers, dimers and tetramers whose relative populations shift to tetramers with the increase of peptide-to-lipid (P/L) molar ratio. Our results strongly support the tandem mechanism of M2 assembly that is monomers-to-dimer then dimers-to-tetramer, since tight dimers are abundant at small P/L’s and thereafter they assemble as dimers of dimers in weaker tetramers. The stepwise mechanism found for a single-pass membrane protein oligomeric assembly should contribute to the knowledge of the association steps in membrane protein folding.
|
|
|
Dynamic Nuclear Polarization of Membrane Proteins: Covalently Bound Spin-Labels at Protein-Protein Interfaces
B.J. Wylie, B.G. Dzikovski, S. Pawsey, M. Caporini, M. Rosay, J.H. Freed, A.E. McDermott.
J. Biomol. NMR 61, 361-367 (2015).
Supporting Information
<doi: 10.1007/s10858-015-9919-6>
PMID:
25828256
PMCID:
PMC4819240
Publication #397
|

|
|
ABSTRACT: We demonstrate that dynamic nuclear polarization of membrane proteins in lipid bilayers may be achieved using a novel polarizing agent: pairs of spin labels covalently bound to a protein of interest interacting at an intermolecular interaction surface. For gramicidin A, nitroxide tags attached to the N-terminal intermolecular interface region become proximal only when bimolecular channels forms in the membrane. We obtained signal enhancements of sixfold for the dimeric protein. The enhancement effect was comparable to that of a doubly tagged sample of gramicidin C, with intramolecular spin pairs. This approach could be a powerful and selective means for signal enhancement in membrane proteins, and for recognizing intermolecular interfaces.
|
|
|
Protein Dynamics in the Solid State from 2H NMR Line Shape Analysis: A Consistent Perspective
E. Meirovitch, Z. Liang, and J.H. Freed.
J. Phys. Chem B 119, 2857-2868 (2015).
<doi: 10.1021/jp511386b>
PMID:
25594631
PMCID:
PMC4358757
Publication #396
|

|
|
ABSTRACT: Deuterium line shape analysis of CD3 groups has emerged as a particularly useful tool for studying microsecond–millisecond protein motions in the solid state. The models devised so far consist of several independently conceived simple jump-type motions. They are comprised of physical quantities encoded in their simplest form; improvements are only possible by adding yet another simple motion, thereby changing the model. The various treatments developed are case-specific; hence comparison among the different systems is not possible. Here we develop a new methodology for 2H NMR line shape analysis free of these limitations. It is based on the microscopic-order-macroscopic-disorder (MOMD) approach. In MOMD motions are described by diffusion tensors, spatial restrictions by potentials/ordering tensors, and geometric features by relative tensor orientations. Jump-type motions are recovered in the limit of large orientational potentials. Model improvement is accomplished by monitoring the magnitude, symmetry, and orientation of the various tensors. The generality of MOMD makes possible comparison among different scenarios. CD3 line shapes from the Chicken Villin Headpiece Subdomain and the Streptomyces Subtilisin Inhibitor are used as experimental examples. All of these spectra are reproduced by using rhombic local potentials constrained for simplicity to be given by the L = 2 spherical harmonics, and by axial diffusion tensors. Potential strength and rhombicity are found to be ca. 2–3 kBT. The diffusion tensor is tilted at 120° from the C–CD3 axis. The perpendicular (parallel) correlation times for local motion are 0.1–1.0 ms (3.3–30 μs). Activation energies in the 1.1–8.0 kcal/mol range are estimated. Future prospects include extension to the 2H relaxation limit, application to the 15N and 13C NMR nuclei, and accounting for collective motions and anisotropic media.
|
|
|
Assembly States of FliM and FliG Within the Flagellar Switch Complex
R. Sircar, P.P. Borbat, M.J. Lynch, J. Bhatnagar, M.S. Beyersdorf, C.J. Halkides, J.H. Freed and B.R. Crane.
J. Mol. Biol. 427, 867-886 (2015).
Supporting Information
<doi: 10.1016/j.jmb.2014.12.009>
PMID:
25536293
PMCID:
PMC4323944
Publication #395
|
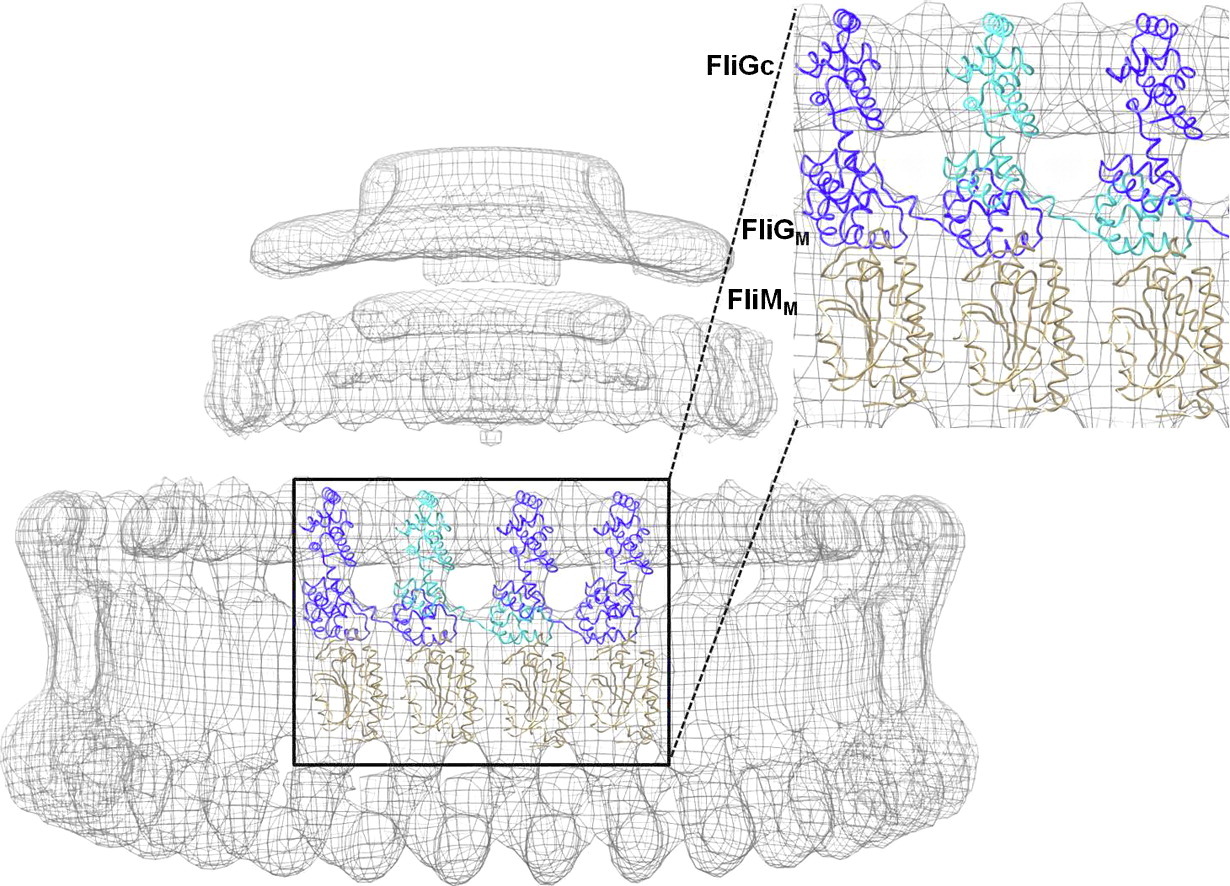
|
|
ABSTRACT: At the base of the bacterial flagella, a cytoplasmic rotor (the C-ring) generates torque and reverses rotation sense in response to stimuli. The bulk of the C-ring forms from many copies of the proteins FliG, FliM, and FliN, which together constitute the switch complex. To help resolve outstanding issues regarding C-ring architecture, we have investigated interactions between FliM and FliG from Thermotoga maritima with X-ray crystallography and pulsed dipolar ESR spectroscopy (PDS). A new crystal structure of an 11-unit FliG:FliM complex produces a large arc with a curvature consistent with the dimensions of the C-ring. Previously determined structures along with this new structure provided a basis to test switch complex assembly models. PDS combined with mutational studies and targeted cross-linking reveal that FliM and FliG interact through their middle domains to form both parallel and antiparallel arrangements in solution. Residue substitutions at predicted interfaces disrupt higher-order complexes that are primarily mediated by contacts between the C-terminal domain of FliG and the middle domain of a neighboring FliG molecule. Spin separations among multi-labeled components fit a self-consistent model that agree well with electron microscopy images of the C-ring. An activated form of the response regulator CheY destabilizes the parallel arrangement of FliM molecules to perturb FliG alignment in a process that may reflect the onset of rotation switching. These data suggest a model of C-ring assembly in which intermolecular contacts among FliG domains provide a template for FliM assembly and cooperative transitions.
|
|
|
Bacterial Chemoreceptor Dynamics Correlate With Activity State and Are Coupled Over Long Distances
D. Samanta, P.P. Borbat, B. Dzikovski, J.H. Freed, and B.R. Crane.
Proc. Natl. Acad. Sci. U. S. A. 112, 2455-2460 (2015).
Supporting Information
<doi: 10.1073/pnas.1414155112>
PMID:
25675479
PMCID:
PMC4345563
Publication #394
|

|
|
SIGNIFICANCE: Bacterial chemoreceptors are a key system for understanding how conformational signals propagate over large distances in transmembrane signaling. We have applied pulsed dipolar ESR spectroscopy of spin-labeled receptors to correlate conformation and dynamics with activity state. We find that the receptor cytoplasmic domain behaves as one large dynamically coupled system, in which activation signals destabilize membrane proximal regions but stabilize the most distal protein interaction tip. Inhibitory signals or adaptations of the receptor through chemical modification produce the opposite changes in conformational properties. This reciprocal coupling of conformational stability provides a versatile mechanism for sending signals throughout large modular proteins.
ABSTRACT: Dynamics are hypothesized to play an important role in the transmission of signals across membranes by receptors. Bacterial chemoreceptors are long helical proteins that consist of a periplasmic ligand-binding domain; a transmembrane region; a cytoplasmic HAMP (histidine kinase, adenylyl cyclases, methyl-accepting chemotaxis proteins, and phosphatases) domain; and a kinase-control module (KCM). The KCM is further composed of adaptation, hinge, and protein interaction regions (PIRs), the latter of which binds the histidine kinase CheA and adaptor CheW. Fusions of the Escherichia coli aspartate receptor KCM to HAMP domains of defined structure (H1-Tar vs. H1-2-Tar) give opposite responses in phosphotransfer and cellular assays, despite similar binding to CheA and CheW. Pulsed dipolar ESR spectroscopy (PDS) of these isolated on and off dimeric effectors reveals that, in the kinase-on state, the HAMP is more conformationally destabilized compared with the PIR, whereas in the kinase-off state, the HAMP is more compact, and the PIR samples a greater breadth of conformations. On and off HAMP states produce different conformational effects at the KCM junction, but these differences decrease through the adaptation region and into the hinge only to return with the inverted relationship in the PIR. Continuous wave–ESR of the spin-labeled proteins confirms that broader PDS distance distributions correlate with increased rates of dynamics. Conformational breadth in the adaptation region changes with charge alterations caused by modification enzymes. Activating modifications broaden the HAMP conformational ensemble but correspondingly, compact the PIR. Thus, chemoreceptors behave as coupled units, in which dynamics in regions proximal and distal to the membrane change coherently but with opposite sign.
|
|
|
Pulse Dipolar ESR of Doubly Labeled Mini TAR DNA and Its Annealing to Mini TAR RNA
Y. Sun, P.P. Borbat, V.M. Grigoryants, W.K. Myers, J.H. Freed, and C.P. Scholes.
Biophys. J. 108, 893-902 (2015).
Supporting Information
<doi: 10.1016/j.bpj.2014.12.028>
PMID:
25692594
PMCID:
PMC4336369
Publication #393
|

|
|
ABSTRACT: Pulse dipolar electron-spin resonance in the form of double electron electron resonance was applied to strategically placed, site-specifically attached pairs of nitroxide spin labels to monitor changes in the mini TAR DNA stem-loop structure brought on by the HIV-1 nucleocapsid protein NCp7. The biophysical structural evidence was at Ångstrom-level resolution under solution conditions not amenable to crystallography or NMR. In the absence of complementary TAR RNA, double labels located in both the upper and the lower stem of mini TAR DNA showed in the presence of NCp7 a broadened distance distribution between the points of attachment, and there was evidence for several conformers. Next, when equimolar amounts of mini TAR DNA and complementary mini TAR RNA were present, NCp7 enhanced the annealing of their stem-loop structures to form duplex DNA-RNA. When duplex TAR DNA-TAR RNA formed, double labels initially located 27.5 Å apart at the 3′- and 5′-termini of the 27-base mini TAR DNA relocated to opposite ends of a 27 bp RNA-DNA duplex with 76.5 Å between labels, a distance which was consistent with the distance between the two labels in a thermally annealed 27-bp TAR DNA-TAR RNA duplex. Different sets of double labels initially located 26–27 Å apart in the mini TAR DNA upper stem, appropriately altered their interlabel distance to ˜35 Å when a 27 bp TAR DNA-TAR RNA duplex formed, where the formation was caused either through NCp7-induced annealing or by thermal annealing. In summary, clear structural evidence was obtained for the fraying and destabilization brought on by NCp7 in its biochemical function as an annealing agent and for the detailed structural change from stem-loop to duplex RNA-DNA when complementary RNA was present.
|
|
|
Transport Domain Unlocking Sets the Uptake Rate of an Aspartate Transporter
N. Akyuz, E.R. Georgieva, Z. Zhou, S. Stolzenberg, M.A. Cuendet, G. Khelashvili, R.B. Altman, D.S. Terry, J.H. Freed, H. Weinstein, O. Boudker, and S.C. Blanchard.
Nature 518, 68-73 (2015).
|

|
|
Transport Domain Unlocking Sets the Uptake Rate of an Aspartate Transporter
N. Akyuz, E.R. Georgieva, Z. Zhou, S. Stolzenberg, M.A. Cuendet, G. Khelashvili, R.B. Altman, D.S. Terry, J.H. Freed, H. Weinstein, O. Boudker, and S.C. Blanchard.
Nature 518, 68-73 (2015).
Supporting Information
<doi: 10.1038/nature14158>
PMID:
25652997
PMCID:
PMC4351760
Publication #392
|

|
|
ABSTRACT: Glutamate transporters terminate neurotransmission by clearing synaptically released glutamate from the extracellular space, allowing repeated rounds of signalling and preventing glutamate-mediated excitotoxicity. Crystallographic studies of a glutamate transporter homologue from the archaeon Pyrococcus horikoshii, GltPh, showed that distinct transport domains translocate substrates into the cytoplasm by moving across the membrane within a central trimerization scaffold. Here we report direct observations of these 'elevator-like' transport domain motions in the context of reconstituted proteoliposomes and physiological ion gradients using single-molecule fluorescence resonance energy transfer (smFRET) imaging. We show that GltPh bearing two mutations introduced to impart characteristics of the human transporter exhibits markedly increased transport domain dynamics, which parallels an increased rate of substrate transport, thereby establishing a direct temporal relationship between transport domain motion and substrate uptake. Crystallographic and computational investigations corroborated these findings by revealing that the 'humanizing' mutations favour structurally 'unlocked' intermediate states in the transport cycle exhibiting increased solvent occupancy at the interface between the transport domain and the trimeric scaffold.
|
|
|
Tau Binds to Lipid Membrane Surfaces via Short Amphipathic Helices Located in Its Microtubule-Binding Repeats
E.R. Georgieva, S. Xiao, P.P. Borbat, J.H. Freed, and D. Eliezer.
Biophys. J. 107, 1441-1452 (2014).
Supporting Information
<doi: 10.1016/j.bpj.2014.07.046>
PMID:
25229151
PMCID:
PMC4167292
Publication #391
|
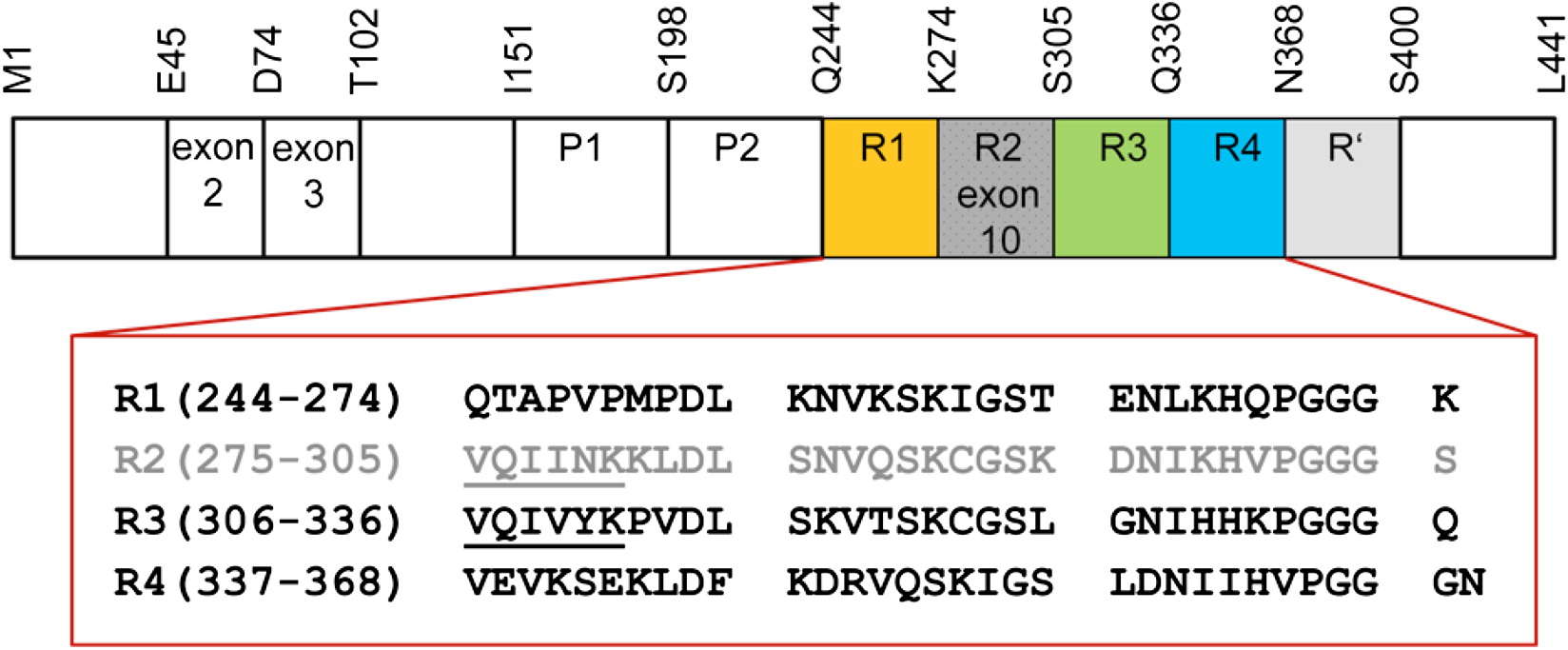
|
|
ABSTRACT: Tau is a microtubule-associated protein that is genetically linked to dementia and linked to Alzheimer's disease via its presence in intraneuronal neurofibrillary tangle deposits, where it takes the form of aggregated paired helical and straight filaments. Although the precise mechanisms by which tau contributes to neurodegeneration remain unclear, tau aggregation is commonly considered to be a critical component of tau-mediated pathogenicity. Nevertheless, the context in which tau aggregation begins in vivo is unknown. Tau is enriched in membrane-rich neuronal structures such as axons and growth cones, and can interact with membranes both via intermediary proteins and directly via its microtubule-binding domain (MBD). Membranes efficiently facilitate tau aggregation in vitro, and may therefore provide a physiologically relevant context for nucleating tau aggregation in vivo. Furthermore, tau-membrane interactions may potentially play a role in tau's poorly understood normal physiological functions. Despite the potential importance of direct tau-membrane interactions for tau pathology and physiology, the structural mechanisms that underlie such interactions remain to be elucidated. Here, we employ electron spin resonance spectroscopy to investigate the secondary and long-range structural properties of the MBD of three-repeat tau isoforms when bound to lipid vesicles and membrane mimetics. We show that the membrane interactions of the tau MBD are mediated by short amphipathic helices formed within each of the MBD repeats in the membrane-bound state. To our knowledge, this is the first detailed elucidation of helical tau structure in the context of intact lipid bilayers. We further show, for the first time (to our knowledge), that these individual helical regions behave as independent membrane-binding sites linked by flexible connecting regions. These results represent the first (to our knowledge) detailed structural view of membrane-bound tau and provide insights into potential mechanisms for membrane-mediated tau aggregation. Furthermore, the results may have implications for the structural basis of tau-microtubule interactions and microtubule-mediated tau aggregation.
|
|
|
Copper-Based Pulsed Dipolar ESR Spectroscopy as a Probe of Protein Conformation Linked to Disease States
G.E. Merz, P.P. Borbat, A.J. Pratt, E.D. Getzoff, J.H. Freed, and B.R. Crane.
Biophys. J. 107, 1669-1674 (2014).
Supporting Information
<doi: 10.1016/j.bpj.2014.07.068>
PMID:
25296320
PMCID:
PMC4190658
Publication #390
|

|
|
ABSTRACT: We demonstrate the ability of pulsed dipolar electron spin resonance (ESR) spectroscopy (PDS) to report on the conformation of Cu-Zn superoxide dismutase (SOD1) through the sensitive measurement of dipolar interactions between inherent Cu2+ ions. Although the extent and the anisotropy of the Cu ESR spectrum provides challenges for PDS, Ku-band (17.3 GHz) double electron-electron resonance and double-quantum coherence variants of PDS coupled with distance reconstruction methods recover Cu-Cu distances in good agreement with crystal structures. Moreover, Cu-PDS measurements expose distinct differences between the conformational properties of wild-type SOD1 and a single-residue variant (I149T) that leads to the disease amyotrophic lateral sclerosis (ALS). The I149T protein displays a broader Cu-Cu distance distribution within the SOD1 dimer compared to wild-type. In a nitroxide (NO)-labeled sample, distance distributions obtained from Cu-Cu, Cu-NO, and NO-NO separations reveal increased structural heterogeneity within the protein and a tendency for mutant dimers to associate. In contrast, perturbations caused by the ALS mutation are completely masked in the crystal structure of I149T. Thus, PDS readily detects alterations in metalloenzyme solution properties not easily deciphered by other methods and in doing so supports the notion that increased range of motion and associations of SOD1 ALS variants contribute to disease progression.
|
|
|
Aggregation Propensities of Superoxide Dismutase G93 Hotspot Mutants Mirror ALS Clinical Phenotypes
A.J. Pratt, D.S. Shin, G.E. Merz, R.P. Rambo, W.A. Lancaster, K.N. Dyer, P.P. Borbat, F.L. Poole II, M.W.W. Adams, J.H. Freed, B.R. Crane, J.A. Tainer, and E.D. Getzoff.
Proc. Natl. Acad. Sci. U. S. A. 111, E4568-E4576 (2014).
|
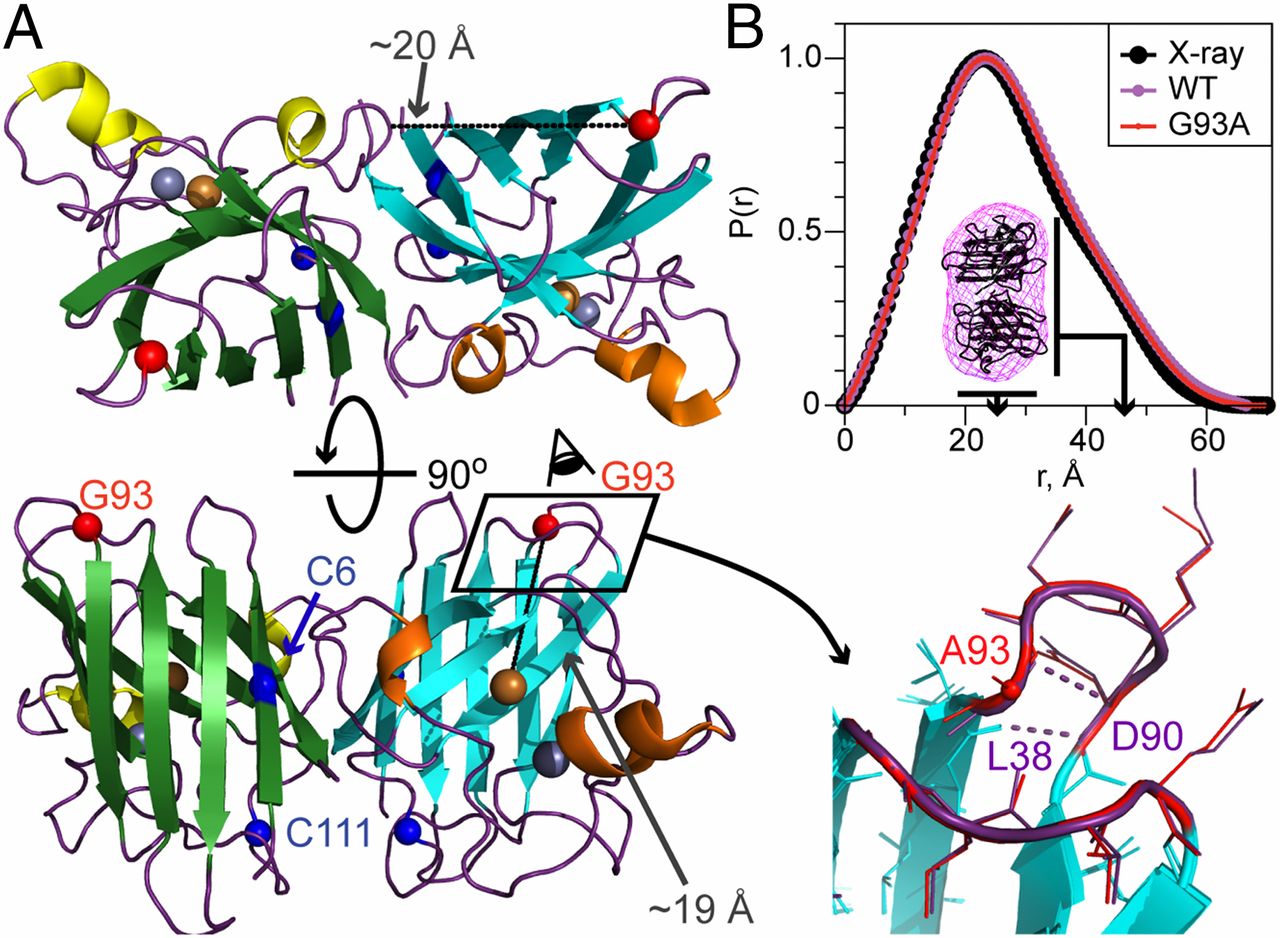
|
|
Aggregation Propensities of Superoxide Dismutase G93 Hotspot Mutants Mirror ALS Clinical Phenotypes
A.J. Pratt, D.S. Shin, G.E. Merz, R.P. Rambo, W.A. Lancaster, K.N. Dyer, P.P. Borbat, F.L. Poole II, M.W.W. Adams, J.H. Freed, B.R. Crane, J.A. Tainer, and E.D. Getzoff.
Proc. Natl. Acad. Sci. U. S. A. 111, E4568-E4576 (2014).
Supporting Information
<doi: 10.1073/pnas.1308531111>
PMID:
25316790
PMCID:
PMC4217430
Publication #389
|

|
|
SIGNIFICANCE: Mutations in human Cu, Zn superoxide dismutase (SOD) cause the motor neuron disease ALS. To better understand why, we compared the aggregation, metal binding, and conformational dynamics of normal and mutant SOD proteins by using the biophysical techniques of X-ray scattering, inductively coupled plasma MS, and ESR spectroscopy. For SOD proteins with defects at a mutational hotspot, we found that copper deficiency, flexibility, and aggregation paralleled clinical severity in ALS patients. These data support a unifying protein framework destabilization mechanism for SOD-linked ALS and thereby point to potential therapies for this lethal condition with few treatment options.
ABSTRACT: Protein framework alterations in heritable Cu, Zn superoxide dismutase (SOD) mutants cause misassembly and aggregation in cells affected by the motor neuron disease ALS. However, the mechanistic relationship between superoxide dismutase 1 (SOD1) mutations and human disease is controversial, with many hypotheses postulated for the propensity of specific SOD mutants to cause ALS. Here, we experimentally identify distinguishing attributes of ALS mutant SOD proteins that correlate with clinical severity by applying solution biophysical techniques to six ALS mutants at human SOD hotspot glycine 93. A small-angle X-ray scattering (SAXS) assay and other structural methods assessed aggregation propensity by defining the size and shape of fibrillar SOD aggregates after mild biochemical perturbations. Inductively coupled plasma MS quantified metal ion binding stoichiometry, and pulsed dipolar ESR spectroscopy evaluated the Cu2+ binding site and defined cross-dimer copper–copper distance distributions. Importantly, we find that copper deficiency in these mutants promotes aggregation in a manner strikingly consistent with their clinical severities. G93 mutants seem to properly incorporate metal ions under physiological conditions when assisted by the copper chaperone but release copper under destabilizing conditions more readily than the WT enzyme. Altered intradimer flexibility in ALS mutants may cause differential metal retention and promote distinct aggregation trends observed for mutant proteins in vitro and in ALS patients. Combined biophysical and structural results test and link copper retention to the framework destabilization hypothesis as a unifying general mechanism for both SOD aggregation and ALS disease progression, with implications for disease severity and therapeutic intervention strategies.
|
|
|
Dph3 Is an Electron Donor for Dph1-Dph2 in the First Step of Eukaryotic Diphthamide Biosynthesis
M. Dong, X. Su, B. Dzikovski, E.E. Dando, X. Zhu, J. Du, J.H. Freed, and H. Lin.
J. Am. Chem. Soc. 136, 1754-1757 (2014).
Supporting Information
<doi: 10.1021/ja4118957>
PMID:
24422557
PMCID:
PMC3985478
Publication #388
|

|
|
ABSTRACT: Diphthamide, the target of diphtheria toxin, is a unique posttranslational modification on translation elongation factor 2 (EF2) in archaea and eukaryotes. The biosynthesis of diphthamide was proposed to involve three steps. The first step is the transfer of the 3-amino-3-carboxypropyl group from S-adenosyl-L-methionine (SAM) to the histidine residue of EF2, forming a C–C bond. Previous genetic studies showed this step requires four proteins in eukaryotes, Dph1–Dph4. However, the exact molecular functions for the four proteins are unknown. Previous study showed that Pyrococcus horikoshii Dph2 (PhDph2), a novel iron-sulfur cluster-containing enzyme, forms a homodimer and is sufficient for the first step of diphthamide biosynthesis in vitro. Here we demonstrate by in vitro reconstitution that yeast Dph1 and Dph2 form a complex (Dph1-Dph2) that is equivalent to the homodimer of PhDph2 and is sufficient to catalyze the first step in vitro in the presence of dithionite as the reductant. We further demonstrate that yeast Dph3 (also known as KTI11), a CSL-type zinc finger protein, can bind iron and in the reduced state can serve as an electron donor to reduce the Fe-S cluster in Dph1-Dph2. Our study thus firmly establishes the functions for three of the proteins involved in eukaryotic diphthamide biosynthesis. For most radical SAM enzymes in bacteria, flavodoxins and flavodoxin reductases are believed to serve as electron donors for the Fe-S clusters. The finding that Dph3 is an electron donor for the Fe-S clusters in Dph1-Dph2 is thus interesting and opens up new avenues of research on electron transfer to Fe-S proteins in eukaryotic cells.
|
|
|
HIV gp41 Fusion Peptide Increases Membrane Ordering in a Cholesterol-Dependent Fashion
A.L. Lai and J.H. Freed.
Biophys. J. 106, 172-181 (2014).
Supporting Information
<doi: 10.1016/j.bpj.2013.11.027>
PMID:
24411249
PMCID:
PMC3907210
Publication #387
|
|
|
ABSTRACT: Fusion between viral envelopes and host cell membranes, which is mediated by special glycoproteins anchored on the viral membrane, is required for HIV viral entry and infection. The HIV gp41 fusion peptide (FP), which initiates membrane fusion, adopts either an α-helical or β-sheeted structure depending on the cholesterol concentration. We used phosphocholine spin labels on the lipid headgroup and different positions on the acyl chain to detect its perturbation on lipid bilayers containing different cholesterol concentrations by electron-spin resonance. Our findings were as follows. 1), gp41 FP affects the lipid order in the same manner as previously shown for influenza hemagglutinin FP, i.e., it has a cooperative effect versus the peptide/lipid ratio, supporting our hypothesis that membrane ordering is a common prerequisite for viral membrane fusion. 2), gp41 FP induces membrane ordering in all lipid compositions studied, whereas a nonfusion mutant FP perturbs lipid order to a significantly smaller extent. 3), In high-cholesterol-containing lipid bilayers, where gp41 FP is in the β-aggregation conformation, its effect on the lipid ordering reaches deeper into the bilayer. The different extent to which the two conformers perturb is correlated with their fusogenicity. The possible role of the two conformers in membrane fusion is discussed.
|
|
|
Improved Sensitivity for Long-Distance Measurements in Biomolecules: Five-Pulse Double Electron-Electron Resonance
P.P. Borbat, E.R. Georgieva, and J.H. Freed.
J. Phys. Chem. Lett. 4, 170-175 (2013).
Supporting Information
<doi: 10.1021/jz301788n>
PMID:
23301118
PMCID:
PMC3538160
Publication #386
|

|
|
ABSTRACT: We describe significantly improved long-distance measurements in biomolecules by use of the new multipulse double electron–electron spin resonance (DEER) illustrated with the example of a five-pulse DEER sequence. In this sequence, an extra pulse at the pump frequency is used compared with standard four-pulse DEER. The position of the extra pulse is fixed relative to the three pulses of the detection sequence. This significantly reduces the effect of nuclear spin-diffusion on the electron-spin phase relaxation, thereby enabling longer dipolar evolution times that are required to measure longer distances. Using spin-labeled T4 lysozyme at a concentration less than 50 μM, as an example, we show that the evolution time increases by a factor of 1.8 in protonated solution and 1.4 in deuterated solution to 8 and 12 μs, respectively, with the potential to increase them further. This enables a significant increase in the measurable distances, improved distance resolution, or both.
|
|
|
Architecture of the Soluble Receptor Aer2 Indicates an In-Line Mechanism for PAS and HAMP Domain Signaling
M.V. Airola, D. Huh, N. Sukomon, J. Widom, R. Sircar, P.P. Borbat, J.H. Freed, K.J. Watts, and B.R. Crane.
J. Mol. Biol. 425, 886-901 (2013).
Supporting Information
<doi: 10.1016/j.jmb.2012.12.011>
PMID:
23274111
PMCID:
PMC3577987
Publication #385
|
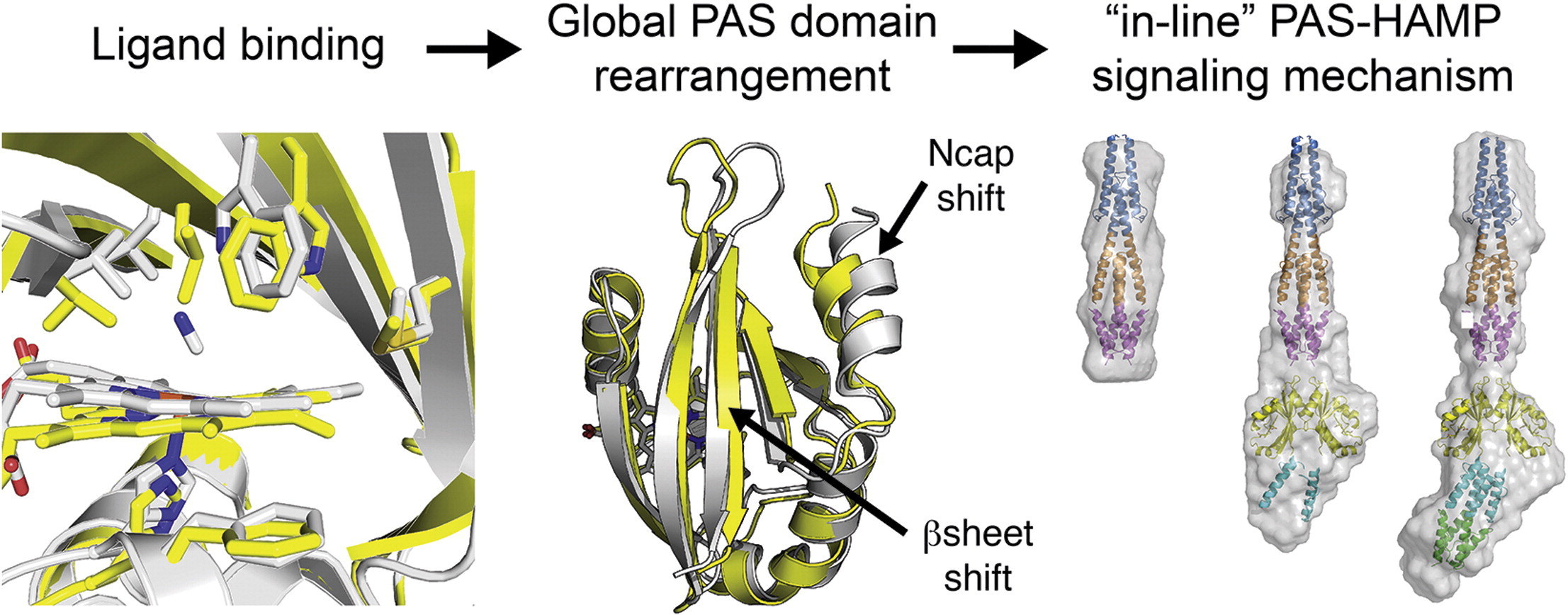
|
|
ABSTRACT: Bacterial receptors typically contain modular architectures with distinct functional domains that combine to send signals in response to stimuli. Although the properties of individual components have been investigated in many contexts, there is little information about how diverse sets of modules work together in full-length receptors. Here, we investigate the architecture of Aer2, a soluble gas-sensing receptor that has emerged as a model for PAS (Per–Arnt–Sim) and poly-HAMP (histidine kinase–adenylyl cyclase–methyl-accepting chemotaxis protein–phosphatase) domain signaling. The crystal structure of the heme-binding PAS domain in the ferric, ligand-free form, in comparison to the previously determined cyanide-bound state, identifies conformational changes induced by ligand binding that are likely essential for the signaling mechanism. Heme-pocket alternations share some similarities with the heme-based PAS sensors FixL and EcDOS but propagate to the Iβ strand in a manner predicted to alter PAS–PAS associations and the downstream HAMP junction within full-length Aer2. Small-angle X-ray scattering of PAS and poly-HAMP domain fragments of increasing complexity allow unambiguous domain assignments and reveal a linear quaternary structure. The Aer2 PAS dimeric crystal structure fits well within ab initio small-angle X-ray scattering molecular envelopes, and pulsed dipolar ESR measurements of inter-PAS distances confirm the crystallographic PAS arrangement within Aer2. Spectroscopic and pull-down assays fail to detect direct interactions between the PAS and HAMP domains. Overall, the Aer2 signaling mechanism differs from the Escherichia coli Aer paradigm, where side-on PAS–HAMP contacts are key. We propose an in-line model for Aer2 signaling, where ligand binding induces alterations in PAS domain structure and subunit association that is relayed through the poly-HAMP junction to downstream domains.
|
|
|
HAMP Domain Conformers that Propagate Opposite Signals in Bacterial Chemoreceptors
M.V. Airola, N. Sukomon, D. Samanta, P.P. Borbat, J.H. Freed, K.J. Watts, and B.R. Crane.
PLoS Biol. 11, e1001479, (2013).
Supporting Information
<doi: 10.1371/journal.pbio.1001479>
PMID:
23424282
PMCID:
PMC3570549
Publication #384
|
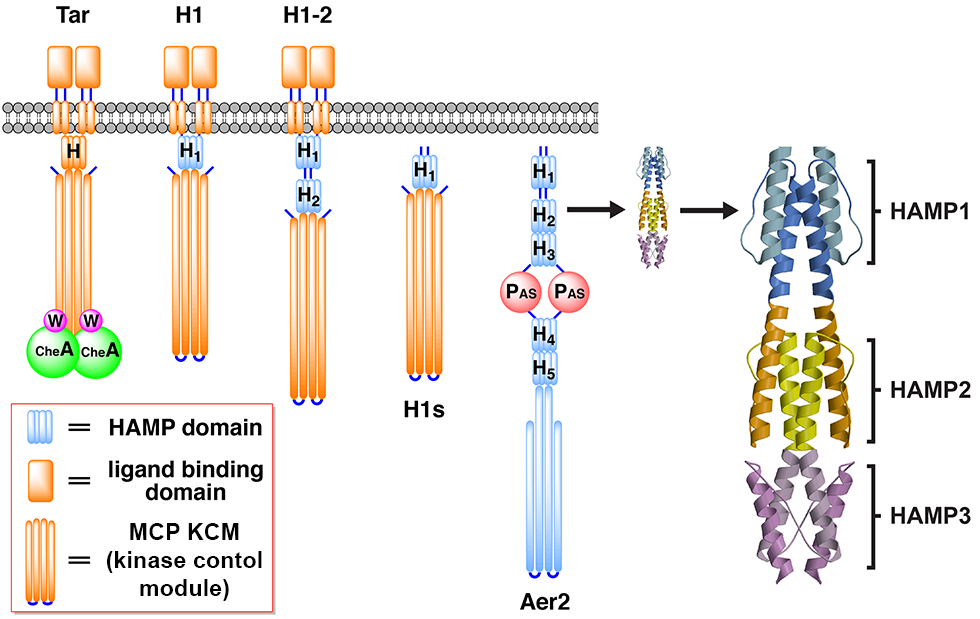
|
|
ABSTRACT: HAMP domains are signal relay modules in >26,000 receptors of bacteria, eukaryotes, and archaea that mediate processes involved in chemotaxis, pathogenesis, and biofilm formation. We identify two HAMP conformations distinguished by a four- to two-helix packing transition at the C-termini that send opposing signals in bacterial chemoreceptors. Crystal structures of signal-locked mutants establish the observed structure-to-function relationships. Pulsed dipolar electron spin resonance spectroscopy of spin-labeled soluble receptors active in cells verify that the crystallographically defined HAMP conformers are maintained in the receptors and influence the structure and activity of downstream domains accordingly. Mutation of HR2, a key residue for setting the HAMP conformation and generating an inhibitory signal, shifts HAMP structure and receptor output to an activating state. Another HR2 variant displays an inverted response with respect to ligand and demonstrates the fine energetic balance between "on" and "off" conformers. A DExG motif found in membrane proximal HAMP domains is shown to be critical for responses to extracellular ligand. Our findings directly correlate in vivo signaling with HAMP structure, stability, and dynamics to establish a comprehensive model for HAMP-mediated signal relay that consolidates existing views on how conformational signals propagate in receptors. Moreover, we have developed a rational means to manipulate HAMP structure and function that may prove useful in the engineering of bacterial taxis responses.
|
|
|
Spin Labels in the Gel Phase and Frozen Lipid Bilayers: Do They Truly Manifest a Polarity Gradient?
B. Dzikowski and J.H. Freed.
In Nitroxides-Theory, Experiment and Applications; A. Kokorin. Ed.
InTech: Rijeka, Croatia, 2012; pp. 167-190, Ch. 5.
<doi: 10.5772/45769>
PMID: [None - Book] PMCID:
[None - Book]
Publication #383
|

|
|
INTRODUCTION: Lipid spin labels containing nitroxide groups at different positions in the fatty acid chain, such as 1-palmitoyl-2-stearoyl-(n-doxyl)-sn-glycero-3-phosphocholines (n-PC spin labels) are a useful and proven tool in lipid research. They have provided important insights into the structure of model and biological membranes, reported on the membrane fluidity, polarity, phase state and presence of microscopic domains, accessibility of different depth positions in the lipid bilayer for oxygen and other polar and non-polar paramagnetic compounds and protein/lipid interactions.
It is generally accepted, that, unlike bulky fluorescent labels, nitroxides are well incorporated into fluid lipid bilayers and not excluded from them. However, it has been shown by NMR that although the most probable location of the nitroxide group for 5-, 10- and 16- PC spin labels in the fluid POPC membrane corresponds to the fully extended conformation, the distribution is relatively broad and other conformations should also be present. Bent conformations were previously found for doxylstearic acids in monomolecular films, water/hydrocarbon emulsion particles and micellar systems. In fluid membranes the fluidity, polarity and accessibility parameters reported by ESR using PC spin labels and n-doxylstearic acids are, in general, change monotonically with an increase in n, although there are indications that the spin label groups on the stearates are located nearer to the membrane exterior than the analogous positions of the unlabeled phospholipid chains. However, in the gel phase, which is characterized by denser chain packing and higher order, the preferential location may be different.
In this chapter we focus on the behavior of PC spin labels in the gel phase and frozen membranes. We show how the superior g-factor resolution of HF ESR provides new insights in this behavior and a new look at the vast body of experimental data accumulated with PC spin labels in the last 30 years. In particular, we revisit so-called "polarity profiles" determined from the g-factor values and hyperfine splittings of PC spin labels in frozen phospholipid membranes with or without cholesterol and show that these values are affected by a number of factors in the membrane composition, chain packing in the lipid phase and folding properties of the sn-2 spin labeled chain PC labels rather than reflect gradients of polarity or water content present in the membrane.
|
|
|
Pulse Dipolar Electron Spin Resonance: Distance Measurements
P.P. Borbat and J.H. Freed.
In Structural Information from Spin-Labels and Intrinsic Paramagnetic Centers in the Biosciences. Structure and Bonding. Vol. 152. J. Harmer and C. Timmel, Eds.
Springer: Heidelberg, Germany; New York, USA, 2013; pp. 1-82.
|

|
|
Pulse Dipolar Electron Spin Resonance: Distance Measurements
P.P. Borbat and J.H. Freed.
In Structural Information from Spin-Labels and Intrinsic Paramagnetic Centers in the Biosciences. Structure and Bonding. Vol. 152. J. Harmer and C. Timmel, Eds.
Springer: Heidelberg, Germany; New York, USA, 2013; pp. 1-82.
<doi: 10.1007/430_2012_82>
PMID: [None - Book] PMCID:
[None - Book]
Publication #382
|

|
|
ABSTRACT: In recent years electron spin resonance (ESR) has provided the means to obtain structural constraints in the field of structural biology on the nanoscale by measuring distances between paramagnetic species, which usually have been nitroxide spin-labels. These ESR methods enable the measurement of distances over the wide range from ca. 6–10 Å to nearly 90 Å. While cw methods may be used for the shortest distances, it is the pulse methods that enable this wide range, as well as determination of the distributions in distance. In this chapter we first describe the underlying theoretical concepts for understanding the principal pulse methods of double quantum coherence (DQC)-ESR and double-electron–electron-resonance (DEER), which we collectively refer to as Pulse-Dipolar ESR Spectroscopies (PDS). We then provide technical aspects of pulse ESR spectrometers required for high quality PDS studies. This is followed by an extensive description of sensitivity considerations in PDS, based largely upon our highly sensitive 17.3 GHz pulse spectrometer at ACERT. This description also includes a comparison of the effectiveness of the respective PDS pulse methods. In addition, the newer methods of 5-pulse DEER, which enables longer distances to be measured than by standard DEER, and 2D-DQC, which provides a convenient mapping for studying orientational coherence between spin labels and their interspin vector, are described.
|
|
|
The Internal Dynamics of Mini c TAR DNA Probed by Electron Paramagnetic Resonance of Nitroxide Spin-Labels at the Lower Stem, the Loop, and the Bulge
Y. Sun, Z. Zhang, V.M. Grigoryants, W.K. Myers, F. Liu, and K. Earle, J.H. Freed, C.P. Scholes
Biochemistry 51, 8530-8541 (2012).
|

|
|
The Internal Dynamics of Mini c TAR DNA Probed by Electron Paramagnetic Resonance of Nitroxide Spin-Labels at the Lower Stem, the Loop, and the Bulge
Y. Sun, Z. Zhang, V.M. Grigoryants, W.K. Myers, F. Liu, and K. Earle, J.H. Freed, C.P. Scholes
Biochemistry 51, 8530-8541 (2012).
Supporting Information
<doi: 10.1021/bi301058q>
PMID:
23009298
PMCID:
PMC3549007
Publication #381
|

|
|
ABSTRACT: Electron paramagnetic resonance (EPR) at 236.6 and 9.5 GHz probed the tumbling of nitroxide spin probes in the lower stem, in the upper loop, and near the bulge of mini c TAR DNA. High-frequency 236.6 GHz EPR, not previously applied to spin-labeled oligonucleotides, was notably sensitive to fast, anisotropic, hindered local rotational motion of the spin probe, occurring approximately about the NO nitroxide axis. Labels attached to the 2′-aminocytidine sugar in the mini c TAR DNA showed such anisotropic motion, which was faster in the lower stem, a region previously thought to be partially melted. More flexible labels attached to phosphorothioates at the end of the lower stem tumbled isotropically in mini c TAR DNA, mini TAR RNA, and ψ3 RNA, but at 5 °C, the motion became more anisotropic for the labeled RNAs, implying more order within the RNA lower stems. As observed by 9.5 GHz EPR, the slowing of nanosecond motions of large segments of the oligonucleotide was enhanced by increasing the ratio of the nucleocapsid protein NCp7 to mini c TAR DNA from 0 to 2. The slowing was most significant at labels in the loop and near the bulge. At a 4:1 ratio of NCp7 to mini c TAR DNA, all labels reported tumbling times of >5 ns, indicating a condensation of NCp7 and TAR DNA. At the 4:1 ratio, pulse dipolar EPR spectroscopy of bilabels attached near the 3′ and 5′ termini showed evidence of an NCp7-induced increase in the 3′–5′ end-to-end distance distribution and a partially melted stem.
|
|
|
Conformational Ensemble of the Sodium-Coupled Aspartate Transporter
E.R. Georgieva, P.P. Borbat, C. Ginter, J.H. Freed, and O. Boudker.
Nat. Struct. Mol. Biol. 20, 215-221 (2013).
Supporting Information
<doi: 10.1038/nsmb.2494>
PMID:
23334289
PMCID:
PMC3565060
Publication #380
|

|
|
ABSTRACT: Sodium and aspartate symporter from Pyrococcus horikoshii, GltPh, is a homolog of the mammalian glutamate transporters, homotrimeric integral membrane proteins that control neurotransmitter levels in brain synapses. These transporters function by alternating between outward-facing and inward-facing states, in which the substrate binding site is oriented toward the extracellular space and the cytoplasm, respectively. Here we used double electron-electron resonance (DEER) spectroscopy to probe the structure and the state distribution of the subunits in the trimer in distinct hydrophobic environments of detergent micelles and lipid bilayers. Our experiments reveal a conformational ensemble of protomers that sample the outward-facing and inward-facing states with nearly equal probabilities, indicative of comparable energies, and independently of each other. On average, the distributions varied only modestly in detergent and in bilayers, but in several mutants unique conformations were stabilized by the latter.
|
|
|
Locating a Lipid at the Portal to the Lipoxygenase Active Site
B.J. Gaffney, M. Bradshaw, S. Frausto, F. Wu, J.H. Freed, and P.P. Borbat.
Biophysical J. 103, 2134-2144, (2012).
Supporting Information
<doi: 10.1016/j.bpj.2012.10.002>
PMID:
23200047
PMCID:
PMC3512035
Publication #379
|
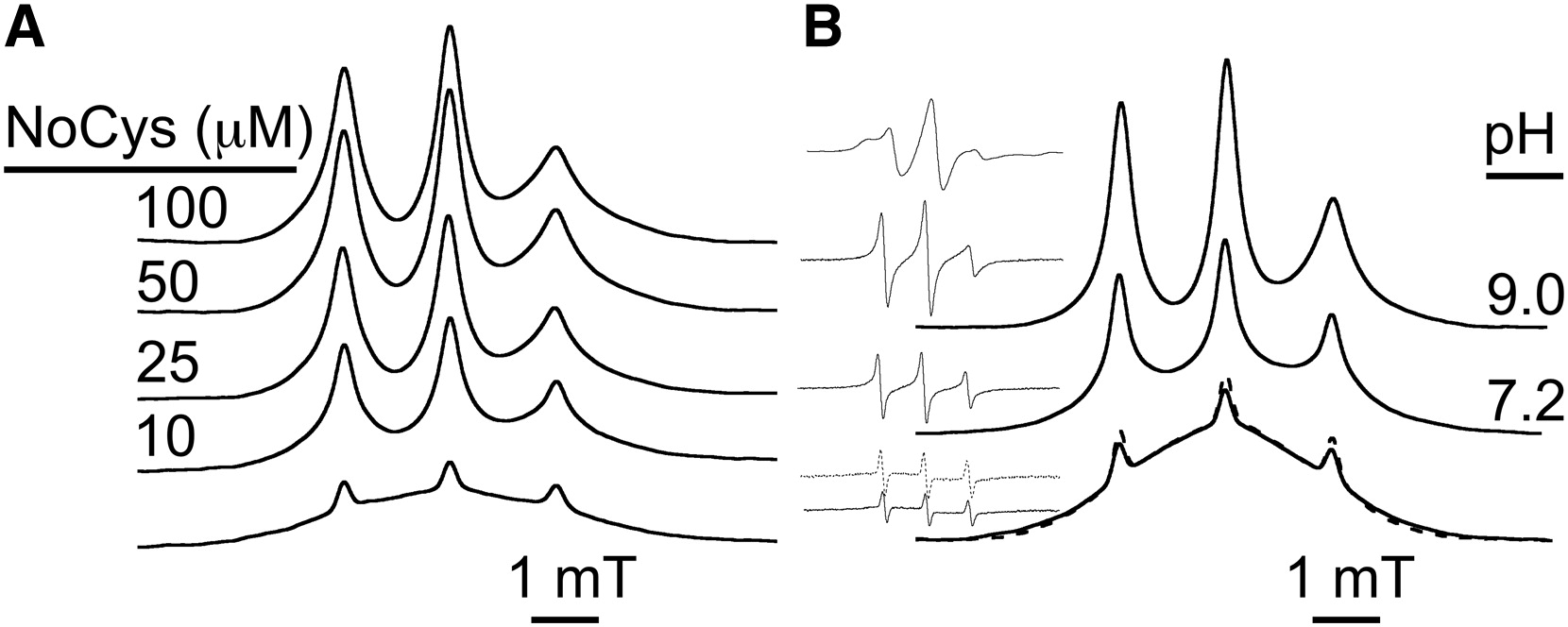
|
|
ABSTRACT: Lipoxygenase enzymes initiate diverse signaling pathways by specifically directing oxygen to different carbons of arachidonate and other polyunsaturated acyl chains, but structural origins of this specificity have remained unclear. We therefore determined the nature of the lipoxygenase interaction with the polar-end of a paramagnetic lipid by electron paramagnetic resonance spectroscopy. Distances between selected grid points on soybean seed lipoxygenase-1 (SBL1) and a lysolecithin spin-labeled on choline were measured by pulsed (electron) dipolar spectroscopy. The protein grid was designed by structure-based modeling so that five natural side chains were replaced with spin labels. Pairwise distances in 10 doubly spin-labeled mutants were examined by pulsed dipolar spectroscopy, and a fit to the model was optimized. Finally, experimental distances between the lysolecithin spin and each single spin site on SBL1 were also obtained. With these 15 distances, distance geometry localized the polar-end and the spin of the lysolecithin to the region between the two domains in the SBL1 structure, nearest to E236, K260, Q264, and Q544. Mutation of a nearby residue, E256A, relieved the high pH requirement for enzyme activity of SBL1 and allowed lipid binding at pH 7.2. This general approach could be used to locate other flexible molecules in macromolecular complexes.
|
|
|
Pulsed ESR Dipolar Spectroscopy for Distance Measurements in Immobilized Spin Labeled Proteins in Liquid Solution
Z. Yang, Y. Liu, P.P. Borbat, J.L. Zweier, J.H. Freed, and W.L. Hubbell.
J. Am. Chem. Soc. 134, 9950-9952 (2012).
Supporting Information
<doi: 10.1021/ja303791p>
PMID:
22676043
PMCID:
PMC3409244
Publication #378
|

|
|
ABSTRACT: Pulsed electron spin resonance (ESR) dipolar spectroscopy (PDS) in combination with site-directed spin labeling is unique in providing nanometer-range distances and distributions in biological systems. To date, most of the pulsed ESR techniques require frozen solutions at cryogenic temperatures to reduce the rapid electron spin relaxation rate and to prevent averaging of electron–electron dipolar interaction due to the rapid molecular tumbling. To enable measurements in liquid solution, we are exploring a triarylmethyl (TAM)-based spin label with a relatively long relaxation time where the protein is immobilized by attachment to a solid support. In this preliminary study, TAM radicals were attached via disulfide linkages to substituted cysteine residues at positions 65 and 80 or 65 and 76 in T4 lysozyme immobilized on Sepharose. Interspin distances determined using double quantum coherence (DQC) in solution are close to those expected from models, and the narrow distance distribution in each case indicates that the TAM-based spin label is relatively localized.
|
|
|
Self-Association of the Histidine Kinase CheA as Studied by Pulsed Dipolar ESR Spectroscopy
J. Bhatnagar, R. Sircar, P.P. Borbat, J.H. Freed, and B.R. Crane.
Biophys. J. 102, 2192-2201 (2012).
Supporting Information
<doi: 10.1016/j.bpj.2012.03.038>
PMID:
22824284
PMCID:
PMC3341552
Publication #377
|

|
|
ABSTRACT: Biologically important protein complexes often involve molecular interactions that are low affinity or transient. We apply pulsed dipolar electron spin resonance spectroscopy and site-directed spin labeling in what to our knowledge is a new approach to study aggregation and to identify regions on protein surfaces that participate in weak, but specific molecular interactions. As a test case, we have probed the self-association of the chemotaxis kinase CheA, which forms signaling clusters with chemoreceptors and the coupling protein CheW at the poles of bacterial cells. By measuring the intermolecular dipolar interactions sensed by spin-labels distributed over the protein surface, we show that the soluble CheA kinase aggregates to a small extent through interactions mediated by its regulatory (P5) domain. Direct dipolar distance measurements confirm that a hydrophobic surface at the periphery of P5 subdomain 2 associates CheA dimers in solution. This result is further supported by differential disulfide cross-linking from engineered cysteine reporter sites. We suggest that the periphery of P5 is an interaction site on CheA for other similar hydrophobic surfaces and plays an important role in structuring the signaling particle.
|
|
|
Dynamics and Ordering of Lipid Spin-Labels Along the Coexistence Curve of Two Membrane Phases: An ESR Study
A.K. Smith and J.H. Freed.
Chem. Phys. Lipids, 165, 348-361 (2012).
Supporting Information
<doi: 10.1016/j.chemphyslip.2012.02.009>
PMID:
22586732
PMCID:
PMC3526980
Publication #376
|

|
|
ABSTRACT: An analysis of electron spin resonance (ESR) spectra from compositions along the liquid-ordered (Lo) and liquid-disordered (Ld) coexistence curve from the brain-sphingomyelin/dioleoylphosphatidylcholine/cholesterol (SPM/DOPC/Chol) model lipid system was performed to characterize the dynamic structure on a molecular level of these coexisting phases. We obtained 200 continuous-wave ESR spectra from glycerophospholipid spin-labels labeled at the 5, 7, 10, 12, 14, and 16 carbon positions of the 2nd acyl chain, a sphingomyelin spin-label labeled at the 14 carbon position of the amide-linked acyl chain, a headgroup-labeled glycerophospholipid, a headgroup-labeled sphingomyelin, and the cholesterol analogue spin-label cholestane all within multi-lamellar vesicle suspensions at room temperature. The spectra were analyzed using the MOMD (microscopic-order macroscopic-disorder) model to provide the rotational diffusion rates and order parameters which characterize the local molecular dynamics in these phases. The analysis also incorporated the known critical point and invariant points of the neighboring three-phase triangle along the coexistence curve. The variation in the molecular dynamic structures of coexisting Lo and Ld compositions as one moves toward the critical point is discussed. Based on these results, a molecular model of the Lo phase is proposed incorporating the "condensing effect" of cholesterol on the phospholipid acyl chain dynamics and ordering and the "umbrella model" of the phospholipid headgroup dynamics and ordering.
|
|
|
Conformational Distributions and Hydrogen Bonding in Gel and Frozen Lipid Bilayers: A High Frequency Spin-Label ESR Study
B. Dzikovski, D. Tipikin, and J. Freed
J. Phys. Chem. B, 116, 6694-6706 (2012).
Supporting Information
<doi: 10.1021/jp211879s>
PMID:
22324811
PMCID:
PMC3376253
Publication #375
|
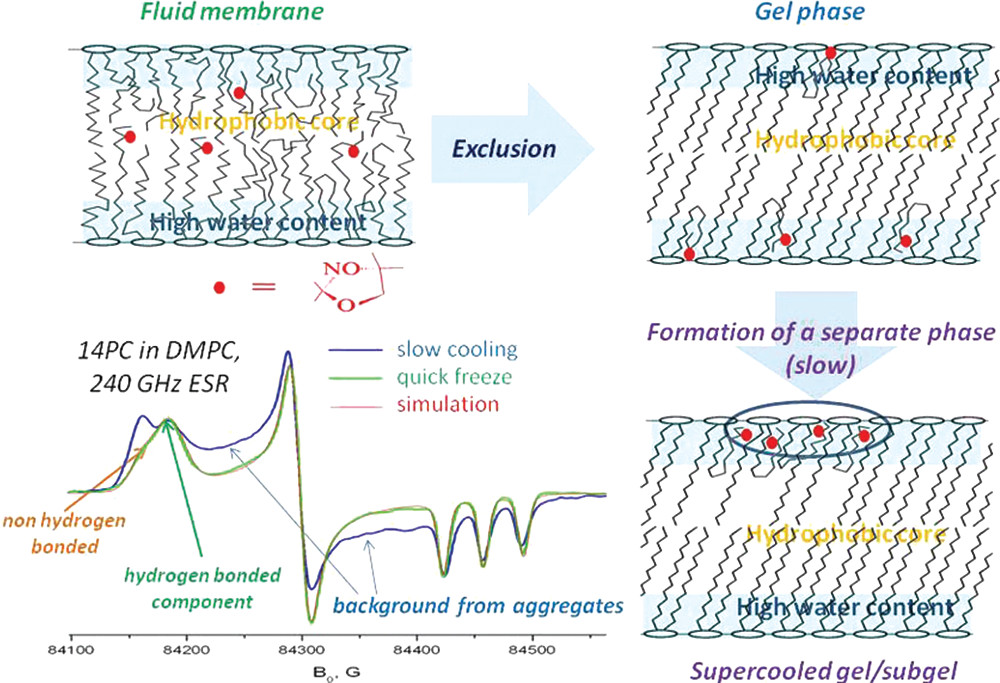
|
|
ABSTRACT: The ESR parameters of PC spin labels in frozen membranes do not simply represent the membrane polarity or water penetration profile. Instead, they show a distribution between hydrogen-bonded (HB) and non-hydrogen-bonded (non-HB) states, which is affected by a number of factors in the membrane composition. Similar to the exclusion of solutes from crystallizing solvents, the pure bulk gel phase excludes nitroxides, forcing acyl chains to take bent conformations. In these conformations, the nitroxide is hydrogen-bonded. Furthermore, upon gradual cooling in the supercooled gel, PC labels undergo slow lateral aggregation, resulting in a broad background signal. However, if the sample is instantly frozen, this background is replaced by the HB component. In membranes with cholesterol, the observed HB/non-HB ratio can best be described by a partition-like equilibrium between nitroxides located in defects of lipid structure within the hydrophobic core and those close to the membrane surface.
|
|
|
Effect of Freezing Conditions on Distances and Their Distributions Derived From Double Electron Electron Resonance (DEER): A Study of Doubly-Spin-Labeled T4 Lysozyme
E.R. Georgieva, A.S. Roy, V.M. Grigoryants, P.P. Borbat, K.A. Earle, C.P. Scholes, and J.H. Freed.
J. Magn. Reson. 216, 69-77 (2012).
Supporting Information
<doi: 10.1016/j.jmr.2012.01.004>
PMID:
22341208
PMCID:
PMC3323113
Publication #374
|

|
|
ABSTRACT: Pulsed dipolar ESR spectroscopy, DEER and DQC, require frozen samples. An important issue in the biological application of this technique is how the freezing rate and concentration of cryoprotectant could possibly affect the conformation of biomacromolecule and/or spin-label. We studied in detail the effect of these experimental variables on the distance distributions obtained by DEER from a series of doubly spin-labeled T4 lysozyme mutants. We found that the rate of sample freezing affects mainly the ensemble of spin-label rotamers, but the distance maxima remain essentially unchanged. This suggests that proteins frozen in a regular manner in liquid nitrogen faithfully maintain the distance-dependent structural properties in solution.
We compared the results from rapidly freeze-quenched (≤100 μs) samples to those from commonly shock-frozen (slow freeze, 1 s or longer) samples. For all the mutants studied we obtained inter-spin distance distributions, which were broader for rapidly frozen samples than for slowly frozen ones. We infer that rapid freezing trapped a larger ensemble of spin label rotamers; whereas, on the time-scale of slower freezing the protein and spin-label achieve a population showing fewer low-energy conformers. We used glycerol as a cryoprotectant in concentrations of 10% and 30% by weight. With 10% glycerol and slow freezing, we observed an increased slope of background signals, which in DEER is related to increased local spin concentration, in this case due to insufficient solvent vitrification, and therefore protein aggregation. This effect was considerably suppressed in slowly frozen samples containing 30% glycerol and rapidly frozen samples containing 10% glycerol. The assignment of bimodal distributions to tether rotamers as opposed to protein conformations is aided by comparing results using MTSL and 4-Bromo MTSL spin-labels. The latter usually produce narrower distance distributions.
|
|
|
A Protocol for Detecting and Scavenging Gas-phase Free Radicals in Mainstream Cigarette Smoke
L.X. Yu, B.G. Dzikovski, and J.H. Freed.
J. Vis. Exp. 59, e3406 (2012).
<doi: 10.3791/3406>
PMID:
22230844
PMCID:
PMC3369772
Publication #373
|

|
|
SUMMARY: Spin-trapping ESR spectroscopy was used to study the effect of plant antioxidants lycopene, pycnogenol and grape seed extract on scavenging gas-phase free radicals in cigarette smoke.
ABSTRACT: Cigarette smoking is associated with human cancers. It has been reported that most of the lung cancer deaths are caused by cigarette smoking. Although tobacco tars and related products in the particle phase of cigarette smoke are major causes of carcinogenic and mutagenic related diseases, cigarette smoke contains significant amounts of free radicals that are also considered as an important group of carcinogens. Free radicals attack cell constituents by damaging protein structure, lipids and DNA sequences and increase the risks of developing various types of cancers. Inhaled radicals produce adducts that contribute to many of the negative health effects of tobacco smoke in the lung. Studies have been conducted to reduce free radicals in cigarette smoke to decrease risks of the smoking-induced damage. It has been reported that haemoglobin and heme-containing compounds could partially scavenge nitric oxide, reactive oxidants and carcinogenic volatile nitrosocompounds of cigarette smoke. A 'bio-filter' consisted of haemoglobin and activated carbon was used to scavenge the free radicals and to remove up to 90% of the free radicals from cigarette smoke. However, due to the cost-ineffectiveness, it has not been successfully commercialized. Another study showed good scavenging efficiency of shikonin, a component of Chinese herbal medicine. In the present study, we report a protocol for introducing common natural antioxidant extracts into the cigarette filter for scavenging gas phase free radicals in cigarette smoke and measurement of the scavenge effect on gas phase free radicals in mainstream cigarette smoke (MCS) using spin-trapping Electron Spin Resonance (ESR) Spectroscopy. We showed high scavenging capacity of lycopene and grape seed extract which could point to their future application in cigarette filters. An important advantage of these prospective scavengers is that they can be obtained in large quantities from byproducts of tomato or wine industry respectively.
|
|
|
Membrane Fluidity
B. Dzikovski and J.H. Freed.
In Encyc. Biophys.; G.C.K. Roberts, Ed.
Springer Verlag: Berlin, Germany, 2013; pp 1440-1446.
|

|
|
Membrane Fluidity
B. Dzikovski and J.H. Freed.
In Encyc. Biophys.; G.C.K. Roberts, Ed.
Springer Verlag: Berlin, Germany, 2013; pp 1440-1446.
<doi: 10.1007/978-3-642-16712-6_546>
PMID: [None - Book] PMCID:
[None - Book]
Publication #372
|

|
|
ABSTRACT: In 1972, Singer and Nicolson (Singer and Nicolson 1972) suggested the so-called fluid mosaic model of the biological membrane (Fig. 1). This useful hypothesis explained many phenomena occurring in model and biological membranes. According to this model, membrane proteins and other membrane-embedded compounds are suspended in a two-dimensional fluid formed by phospholipids. This fluid state of membrane lipids is critical for membrane function. It allows, for example, free diffusion and equal distribution of new cell-synthesized lipids and proteins; lateral diffusion of proteins and other molecules in signaling events and other membrane reactions; membrane fusion, that is, fusion of vesicles with organelles; separation of membranes during cell division; etc.
Membrane fluidity, which describes the ease of movement for molecules in the membrane environment, is a general concept that lacks a precise definition. It is much broader than the strict physical definition of fluidity as the reciprocal of viscosity in the case of isotropic liquids. In general, "membrane fluidity" implies various anisotropic motions, which contribute to the mobility of components of a membrane.
The lipid membrane, as a whole, shows a unique combination of fluidity and rigidity. In terms of the solubility and diffusion of small nonpolar molecules, the membrane behaves very much like an oil drop. In contrast, the translational diffusion constants of lipids and proteins in membranes are characteristic of media with the viscosity over two orders of magnitude greater than that of oil such as hexadecane. Also, in most cases, the membrane represents an impermeable barrier for ions and other hydrophilic compounds.
Figure 2 shows characteristic frequencies (reciprocal of characteristic times) of different kinds of molecular motions in the membrane in comparison to frequency ranges in which various spectroscopic techniques are sensitive to molecular motion (Gennis 1989).
|
|
|
Protein Dynamics by NMR Spin Relaxation: The Slowly Relaxing Local Structure Perspective
E. Meirovitch, A. Polimeno, and J.H. Freed.
In E. Mag. Res. Online, R.K. Harris and R.L. Wasylishen, Eds.
John Wiley & Sons, Inc.: New York, 2011; pp 1-9.
<doi: 10.1002/9780470034590.emrstm1243>
PMID: [None - Book] PMCID:
[None - Book]
Publication #371
|
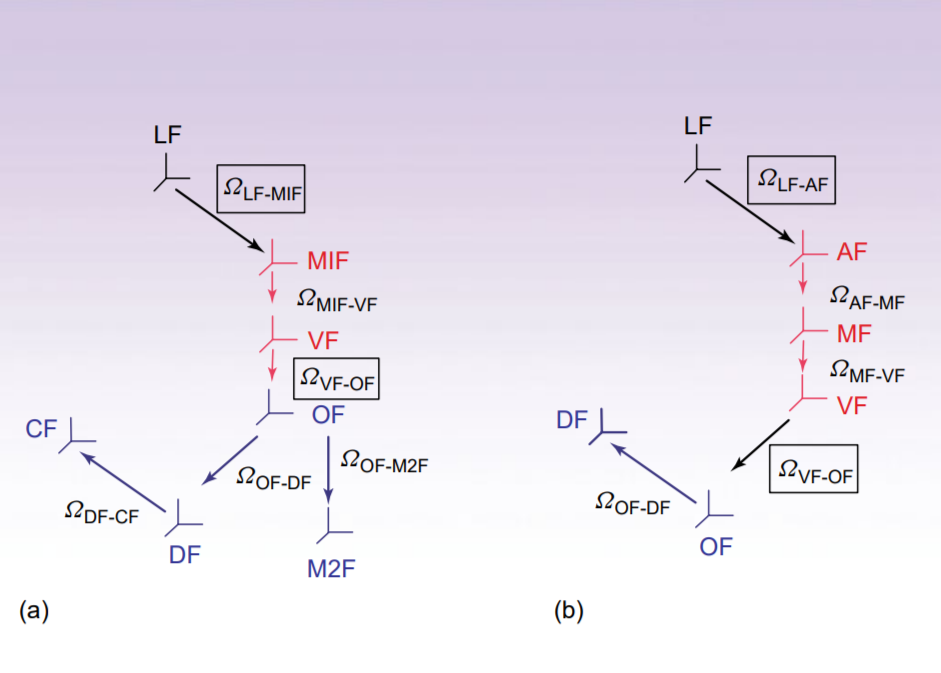
|
|
ABSTRACT: The two-body (protein and probe) coupled rotator slowly relaxing local structure (SRLS) approach, implemented for NMR spin relaxation in proteins, is presented. SRLS is based on a Smoluchowki equation that includes the tensorial descriptions of the diffusing rotators and (via the local ordering) their dynamical coupling. This yields spectral densities that enter the expressions for the experimentally measured relaxation parameters. The constrained motion of the probe is treated by analogy with the standard treatments of restricted motions in liquids. Such restricted motions are largely recovered in the limit of large time scale separation between the rotators, i.e., when mode-coupling is not important. We found that the tensorial properties are not simple for proteins even in this limit. Their complexity has to be accounted for to obtain physically insightful pictures of protein dynamics. The traditional method for analyzing NMR spin relaxation in proteins is model-free (MF). This method treats only the simplest tensorial properties and implies large time-scale separation. Only when these conditions are fulfilled the analytical MF spectral densities, which are limiting cases of SRLS, may be used. Since they are not fulfilled in most actual cases, the MF-based pictures of protein dynamics are unreliable.
|
|
|
Distance Measurements: Continuous-Wave (CW)- and Pulsed Dipolar EPR
S.K. Misra and J.H. Freed.
In Multifrequency Electron Paramagnetic Resonance, S.K. Misra, Ed.
Wiley-VCH: New York, 2011; Chapter 12, pp. 545-588.
<doi: 10.1002/9783527633531.ch12>
PMID: [None - Book] PMCID:
[None - Book]
Publication #370
|
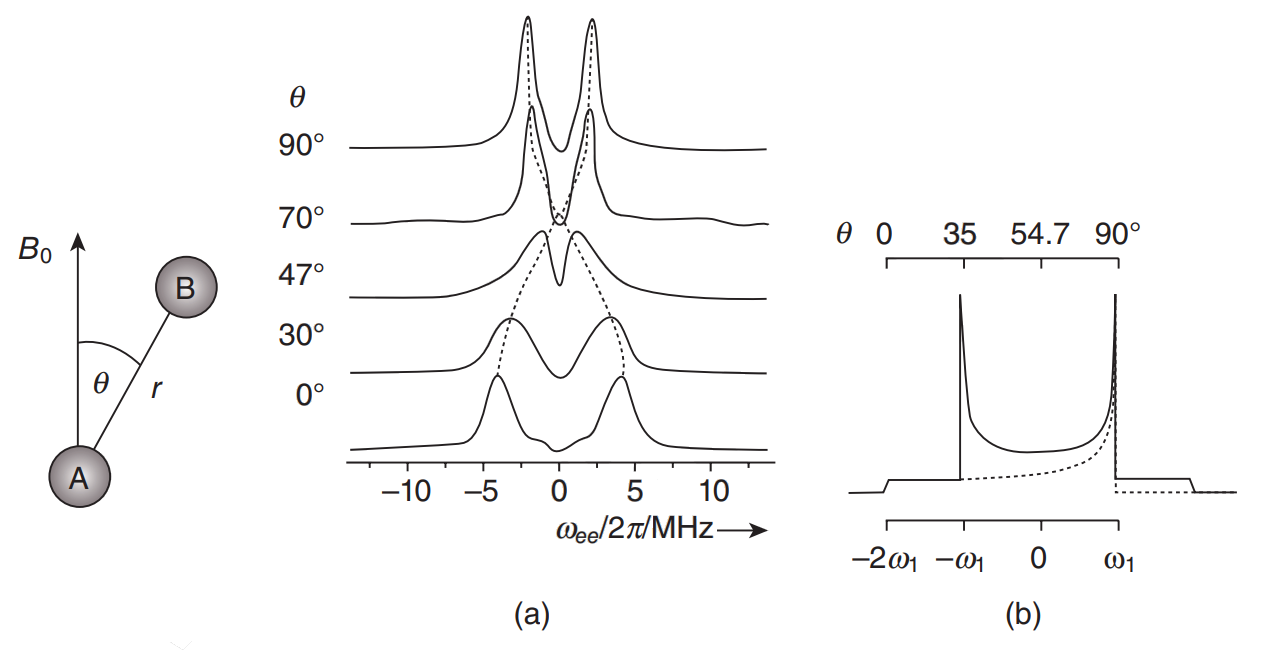
|
|
INTRODUCTION: Information on long-range distances between selected sites is a prerequisite to understanding the detailed structure of complex systems. In NMR utilizing a spin label, it is possible to measure distances in the range between 8 Å to, at most, 25 Å. On the other hand, with EPR, distances in the range between about 10 and 90 Å can and have been measured. EPR spectroscopy may be the best practical technique for complex systems, since methods such as small-angle scattering, X-ray scattering and small-angle neutron scattering have insufficient contrast whereas fluorescence resonant energy transfer (FRET) lacks key virtues of EPR noted below. Distance measurements in EPR are based on exploitation of the dipolar interaction (DI), that depends on the distance between two paramagnetic probes, and which may be identical or may differ from each other, and are located at different spatial positions in the sample. They can be introduced using site-directed mutagenesis (also known as site-directed spin labeling; SDSL), a technique which enables the investigation of, for example, proteins and other biomolecules. In fact, the signature of the DI can be clearly seen in CW-EPR spectra under favorable circumstances for short distances. On the other hand, in pulsed EPR [hereafter referred to as pulsed dipolar spectroscopy; PDS), experiments can be designed specifically to clearly distinguish the dipolar interaction. The two commonly used PDS techniques are: (i) pulsed electron double resonance (PELDOR; this term was originally coined by the Russians, who first developed the technique), which is also referred to as double electron-electron resonance (DEER; this term was introduced later in the USA, and will be used hereafter in this chapter); and (ii) double quantum coherence (DQC)-EPR.
|
|
|
Molecular Motions
S.K. Misra and J.H. Freed.
In Multifrequency Electron Paramagnetic Resonance, S.K. Misra, Ed.
Wiley-VCH: New York, 2011; Chapter 11, pp. 497-544.
|

|
|
Molecular Motions
S.K. Misra and J.H. Freed.
In Multifrequency Electron Paramagnetic Resonance, S.K. Misra, Ed.
Wiley-VCH: New York, 2011; Chapter 11, pp. 497-544.
<doi: 10.1002/9783527633531.ch11>
PMID: [None - Book] PMCID:
[None - Book]
Publication #369
|

|
|
INTRODUCTION: In this chapter we will discuss the study of molecular motion by EPR, using both continuous-wave (CW) and pulsed EPR, in particular two-dimensional electron–electron double resonance (2-D-ELDOR). In recent years, the study of protein dynamics using site-directed spin labels (SDSL) by EPR has become an important subject.
|
|
|
2D-ELDOR Study of Heterogeneity and Domain Structure Changes in Plasma Membrane Vesicles upon Cross-Linking of Receptors
Y.-W. Chiang, A.J. Costa-Filho, B. Baird, and J.H. Freed.
J. Phys. Chem. B 115, 10462-10469 (2011).
Supporting Information
<doi: 10.1021/jp2016243>
PMID:
21780815
PMCID:
PMC3165081
Publication #368
|
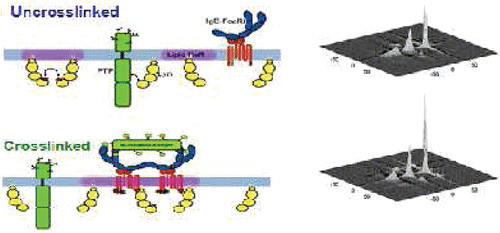
|
|
ABSTRACT: 2D electron–electron double resonance (2D-ELDOR) with the "full Sc–" method of analysis is applied to the study of plasma membrane vesicles. Membrane structural changes upon antigen cross-linking of IgE receptors (IgE-FcεRI) in plasma membrane vesicles (PMVs) isolated from RBL-2H3 mast cells are investigated, for the first time, by means of these 2D-ELDOR techniques. Spectra of 1-palmitoyl-2-(16-doxyl stearoyl) phosphatidylcholine (16-PC) from PMVs before and after this stimulation at several temperatures are reported. The results demonstrate a coexistence of liquid-ordered (Lo) and liquid-disordered (Ld) components. We find that upon cross-linking, the membrane environment is remodeled to become more disordered, as shown by a moderate increase in the population of the Ld component. This change in the relative amount of the Lo versus Ld components upon cross-linking is consistent with a model wherein the IgE receptors, which when clustered by antigen to cause cell stimulation, lead to more disordered lipids, and their dynamic and structural properties are slightly altered. This study demonstrates that 2D-ELDOR, analyzed by the full Sc– method, is a powerful approach for capturing the molecular dynamics in biological membranes. This is a particular case showing how 2D-ELDOR can be applied to study physical processes in complex systems that yield subtle changes.
|
|
|
Electron Tunneling in Lithium-Ammonia Solutions Probed by Frequency-Dependent Electron Spin Relaxation Studies
K. Maeda, M.T.J. Lodge, J. Harmer, J.H. Freed, and P.P. Edwards.
J. Am. Chem. Soc. 134, 9209-18 (2012).
<doi: 10.1021/ja212015b>
PMID:
22568866
PMCID:
PMC3415590
Publication #367
|

|
|
ABSTRACT: Electron transfer or quantum tunneling dynamics for excess or solvated electrons in dilute lithium–ammonia solutions have been studied by pulse electron paramagnetic resonance (EPR) spectroscopy at both X- (9.7 GHz) and W-band (94 GHz) frequencies. The electron spin–lattice (T1) and spin–spin (T2) relaxation data indicate an extremely fast transfer or quantum tunneling rate of the solvated electron in these solutions which serves to modulate the hyperfine (Fermi-contact) interaction with nitrogen nuclei in the solvation shells of ammonia molecules surrounding the localized, solvated electron. The donor and acceptor states of the solvated electron in these solutions are the initial and final electron solvation sites found before, and after, the transfer or tunneling process. To interpret and model our electron spin relaxation data from the two observation EPR frequencies requires a consideration of a multiexponential correlation function. The electron transfer or tunneling process that we monitor through the correlation time of the nitrogen Fermi-contact interaction has a time scale of (1–10) × 10–12 s over a temperature range 230–290 K in our most dilute solution of lithium in ammonia. Two types of electron–solvent interaction mechanisms are proposed to account for our experimental findings. The dominant electron spin relaxation mechanism results from an electron tunneling process characterized by a variable donor–acceptor distance or range (consistent with such a rapidly fluctuating liquid structure) in which the solvent shell that ultimately accepts the transferring electron is formed from random, thermal fluctuations of the liquid structure in, and around, a natural hole or Bjerrum-like defect vacancy in the liquid. Following transfer and capture of the tunneling electron, further solvent-cage relaxation with a time scale of ˜10–13 s results in a minor contribution to the electron spin relaxation times. This investigation illustrates the great potential of multifrequency EPR measurements to interrogate the microscopic nature and dynamics of ultrafast electron transfer or quantum-tunneling processes in liquids. Our results also impact on the universal issue of the role of a host solvent (or host matrix, e.g. a semiconductor) in mediating long-range electron transfer processes and we discuss the implications of our results with a range of other materials and systems exhibiting the phenomenon of electron transfer.
|
|
|
Backbone Dynamics of Deoxy and Carbonmonoxy Hemoglobin by NMR/SRLS
E. Meirovitch, M. Zerbetto, A. Polimeno, and J.H. Freed.
J. Phys. Chem. B 115, 143-157 (2011).
<doi: 10.1021/jp107553j>
PMID:
21162544
PMCID:
PMC3071157
Publication #366
|
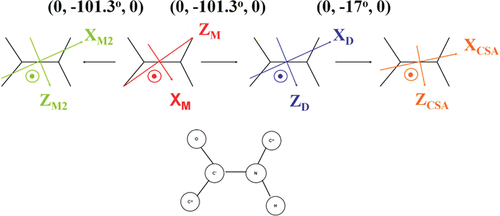
|
|
ABSTRACT: The slowly relaxing local structure (SRLS) approach, developed for NMR spin relaxation analysis in proteins, is applied herein to amide 15N relaxation in deoxy and carbonmonoxy hemoglobin. Experimental data including 15N T1, T2 and 15N-{1H} NOE, acquired at 11.7 and 14.1 T, and 29 and 34 °C, are analyzed. The restricted local motion of the N–H bond is described in terms of the principal value (S02) and orientation (βD) of an axial local ordering tensor, S, and the principal values (R∥L andR⊥L) and orientation (βO) of an axial local diffusion tensor, RL. The parameters c02 (the potential coefficient in terms of which S02 is defined), R∥L, βD, and βO are determined by data fitting; R⊥L is set equal to the global motional rate, RC, found previously to be (5.2–5.8) × 106 1/s in the temperature range investigated. The principal axis of S is (nearly) parallel to the Ci–1α–Ciα axis; when the two axes are parallel, βD = –101.3° (in the frame used). The principal axis of RL is (nearly) parallel to the N–H bond; when the two axes are parallel, βO = –101.3°. For "rigid" N–H bonds located in secondary structure elements the best-fit parameters are S02 = 0.88–0.95 (corresponding to local potentials of 8.6–19.9 kBT), R∥L = 109–1010 1/s, βD = –101.3° ± 2.0°, and βO = –101.3° ± 4°. For flexible N–H bonds located in loops the best-fit values are S02 = 0.75–0.80 (corresponding to local potentials of 4.5–5.5 kBT), R∥L = (1.0–6.3) × 108 1/s, βD = –101.3° ± 4.0°, and βO = –101.3° ± 10°. These results are important in view of their physical clarity, inherent potential for further interpretation, consistency, and new qualitative insights provided (vide infra).
|
|
|
Joint Analysis of ESR Lineshapes and 1H NMRD Profiles of DOTA-Gd Derivatives by Means of the Slow Motion Theory
D. Kruk, J. Kowalewski, D.S. Tipikin, J.H. Freed, M. Mościcki, A. Mielczarek, and M. Port.
J. Chem. Phys. 134, 024508 (2011).
<doi: 10.1063/1.3516590>
PMID:
21241121
PMCID:
PMC3188623
Publication #365
|
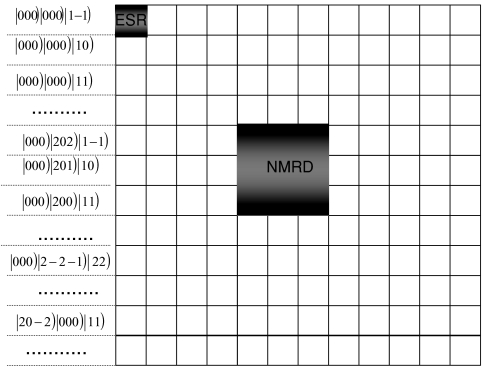
|
|
ABSTRACT: The "Swedish slow motion theory" applied so far to Nuclear Magnetic Relaxation Dispersion (NMRD) profiles for solutions of transition metal ion complexes has been extended to ESR spectral analysis, including in addition g-tensor anisotropy effects. The extended theory has been applied to interpret in a consistent way (within one set of parameters) NMRD profiles and ESR spectra at 95 and 237 GHz for two Gd(III) complexes denoted as P760 and P792 (hydrophilic derivatives of DOTA-Gd, with molecular masses of 5.6 and 6.5 kDa, respectively). The goal is to verify the applicability of the commonly used pseudorotational model of the transient zero field splitting (ZFS). According to this model the transient ZFS is described by a tensor of a constant amplitude, defined in its own principal axes system, which changes its orientation with respect to the laboratory frame according to the isotropic diffusion equation with a characteristic time constant (correlation time) reflecting the time scale of the distortional motion. This unified interpretation of the ESR and NMRD leads to reasonable agreement with the experimental data, indicating that the pseudorotational model indeed captures the essential features of the electron spin dynamics.
|
|
|
Mechanistic Understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] Enzyme Required for Diphthamide Biosynthesis
X. Zhu, B. Dzikovski, X. Su, A.T. Torelli, Y. Zhang, S.E. Ealick, J.H. Freed and H. Lin.
Mol. Biosyst. 7, 74-81 (2011).
<doi: 10.1039/c0mb00076k>
PMID:
20931132
PMCID:
PMC3066188
Publication #364
|

|
|
ABSTRACT: Diphthamide, the target of diphtheria toxin, is a unique posttranslational modification on eukaryotic and archaeal translation elongation factor 2 (EF2). The proposed biosynthesis of diphthamide involves three steps and we have recently found that in Pyrococcus horikoshii (P. horikoshii), the first step uses an S-adenosyl-L-methionine (SAM)-dependent [4Fe–4S] enzyme, PhDph2, to catalyze the formation of a C–C bond. Crystal structure shows that PhDph2 is a homodimer and each monomer contains three conserved cysteine residues that can bind a [4Fe–4S] cluster. In the reduced state, the [4Fe–4S] cluster can provide one electron to reductively cleave the bound SAM molecule. However, different from classical radical SAM family of enzymes, biochemical evidence suggest that a 3-amino-3-carboxypropyl radical is generated in PhDph2. Here we present evidence supporting that the 3-amino-3-carboxypropyl radical does not undergo hydrogen abstraction reaction, which is observed for the deoxyadenosyl radical in classical radical SAM enzymes. Instead, the 3-amino-3-carboxypropyl radical is added to the imidazole ring in the pathway towards the formation of the product. Furthermore, our data suggest that the chemistry requires only one [4Fe–4S] cluster to be present in the PhDph2 dimer.
|
|
|
A New Lanczos-Based Algorithm for Simulating High-Frequency Two-Dimensional Electron Spin Resonance Spectra
Y.-W. Chiang and J.H. Freed.
J. Chem. Phys. 134, 034112 (2011).
<doi: 10.1063/1.3523576>
PMID:
21261335
PMCID:
PMC3041155
Publication #363
|
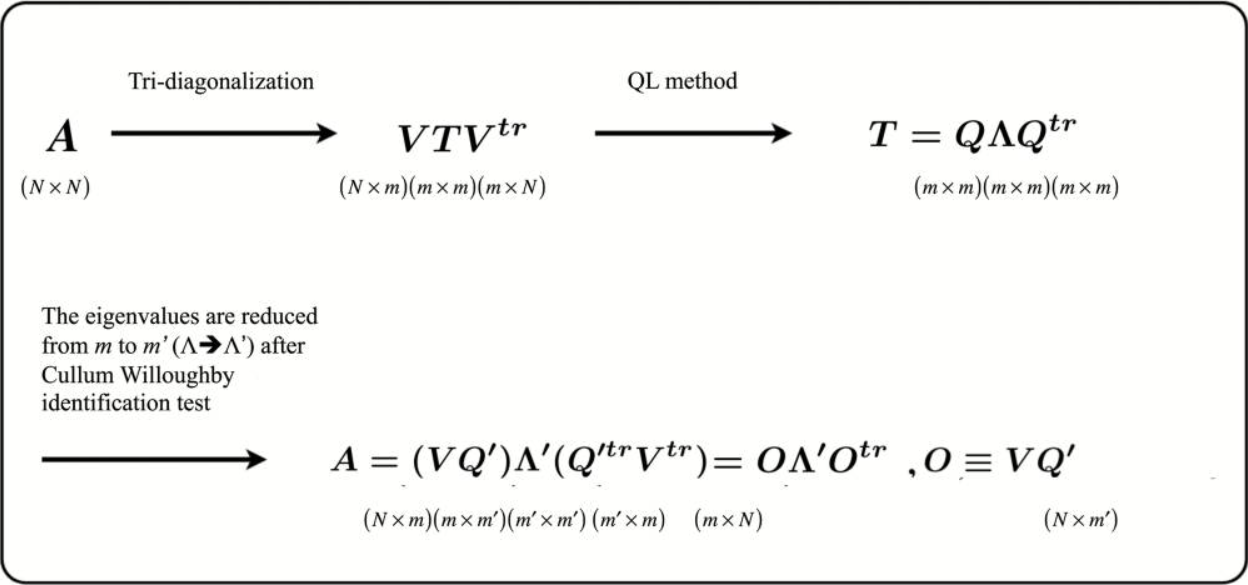
|
|
ABSTRACT: The Lanczos algorithm (LA) is a useful iterative method for the reduction of a large matrix to tridiagonal form. It is a storage efficient procedure requiring only the preceding two Lanczos vectors to compute the next. The quasi-minimal residual (QMR) method is a powerful method for the solution of linear equation systems, Ax = b. In this report we provide another application of the QMR method: we incorporate QMR into the LA to monitor the convergence of the Lanczos projections in the reduction of large sparse matrices. We demonstrate that the combined approach of the LA and QMR can be utilized efficiently for the orthogonal transformation of large, but sparse, complex, symmetric matrices, such as are encountered in the simulation of slow-motional 1D- and 2D-electron spin resonance (ESR) spectra. Especially in the 2D-ESR simulations, it is essential that we store all of the Lanczos vectors obtained in the course of the LA recursions and maintain their orthogonality. In the LA-QMR application, the QMR weight matrix mitigates the problem that the Lanczos vectors lose orthogonality after many LA projections. This enables substantially more Lanczos projections, as required to achieve convergence for the more challenging ESR simulations. It, therefore, provides better accuracy for the eigenvectors and the eigenvalues of the large sparse matrices originating in 2D-ESR simulations than does the previously employed method, which is a combined approach of the LA and the conjugate-gradient (CG) methods, as evidenced by the quality and convergence of the 2D-ESR simulations. Our results show that very slow-motional 2D-ESR spectra at W-band (95 GHz) can be reliably simulated using the LA-QMR method, whereas the LA-CG consistently fails. The improvements due to the LA-QMR are of critical importance in enabling the simulation of high-frequency 2D-ESR spectra, which are characterized by their very high resolution to molecular orientation.
|
|
|
Two Conserved Residues Are Important for Inducing Highly Ordered Membrane Domains by the Transmembrane Domain of Influenza Hemagglutinin
M. Ge and J.H. Freed.
Biophys. J. 100, 90-97 (2011).
Supporting Information
<doi: 10.1016/j.bpj.2010.11.014>
PMID:
21190660
PMCID:
PMC3010018
Publication #362
|

|
|
ABSTRACT: The interaction with lipids of a synthetic peptide corresponding to the transmembrane domain of influenza hemagglutinin was investigated by means of electron spin resonance. A detailed analysis of the electron spin resonance spectra from spin-labeled phospholipids revealed that the major effect of the peptide on the dynamic membrane structure is to induce highly ordered membrane domains that are associated with electrostatic interactions between the peptide and negatively charged lipids. Two highly conserved residues in the peptide were identified as being important for the membrane ordering effect. Aggregation of large unilamellar vesicles induced by the peptide was also found to be correlated with the membrane ordering effect of the peptide, indicating that an increase in membrane ordering, i.e., membrane dehydration, is important for vesicle aggregation. The possibility that hydrophobic interaction between the highly ordered membrane domains plays a role in vesicle aggregation and viral fusion is discussed.
|
|
|
Methyl Dynamics of a Ca2+-Calmodulin-Peptide Complex from NMR/SRLS
Y.E. Shapiro, A. Polimeno, J.H. Freed, and E. Meirovitch.
J. Phys. Chem. B 115, 354-365 (2011).
<doi: 10.1021/jp107130m>
PMID:
21166433
PMCID:
PMC3062514
Publication #361
|

|
|
ABSTRACT: We developed the slowly relaxing local structure (SRLS) approach for analyzing NMR spin relaxation in proteins. SRLS accounts for dynamical coupling between the tumbling of the protein and the local motion of the probe and for general tensorial properties. It is the generalization of the traditional model-free (MF) method, which does not account for mode-coupling and treats only simple tensorial properties. SRLS is applied herein to 2H relaxation of 13CDH2 groups in the complex of Ca2+–calmodulin with the peptide smMLCKp. Literature data comprising 2H T1 and T2 acquired at 14.1 and 17.6 T, and 288, 295, 308, and 320 K, are used. We find that mode-coupling is a small effect for methyl dynamics. On the other hand, general tensorial properties are important. In particular, it is important to allow for the asymmetry of the local spatial restrictions, which can be represented in SRLS by a rhombic local ordering tensor with components S02 and S22. The principal axes frame of this tensor is obviously different from the axial frames of the magnetic tensors. Here, we find that −0.2 ≤ S02 ≤ 0.5 and −0.4 ≤ S22 ≤ 0. MF features a single "generalized" order parameter, S, confined to the 0−0.316 range; the local geometry is inherently simple. The parameter S is inaccurate, having absorbed unaccounted for effects, notably S22 ≠ 0. We find that the methionine methyls (the other methyl types) reorient with rates of 8.6 × 109 to 21.4 × 109 (0.67 × 109 to 6.5 × 109) 1/s. The corresponding activation energies are 10 (10−27) kJ/mol. By contrast, MF yields inaccurate effective local motional correlation times, τe, with nonphysical temperature dependence. Thus, the problematic S- and τe-based MF picture of methyl dynamics has been replaced with an insightful physical picture based on a local ordering tensor related to structural features, and a local diffusion tensor that yields accurate activation energies.
|
|
|
ESR Microscopy for Biological and Biomedical Applications
C.S. Shin, C.R. Dunnam, P.P. Borbat, B. Dzikovski, E.D. Barth, H.J. Halpern, and J.H. Freed.
Nanoscience & Nanotechnology Letters 3, 561-567 (2011).
<doi: 10.1166/nnl.2011.1206>
PMID:
21984955
PMCID:
PMC3188420
Publication #360
|
|
|
ABSTRACT: We report on electron-spin resonance microscopy (ESRM) providing sub-micron resolution (˜700nm) with a high spin concentration sample, i.e. lithium phthalocyanine (LiPc) crystal. For biomedical applications of our ESRM, we have imaged samples containing rat basophilic leukemia (RBL) cells as well as cancerous tissue samples with a resolution of several microns using a water soluble spin probe, Trityl_OX063_d24. Phantom samples with the nitroxide spin label, 15N PDT, were also imaged to demonstrate that nitroxides, which are commonly used as spin labels, may also be used for ESRM applications. ESRM tissue imaging would therefore be valuable for diagnostic or therapeutic purposes. Also, ESRM can be used to study the motility or the metabolism of cells in various environments. With further modification and/or improvement of imaging probe and spectrometer instrumentation sub-micron biological images should be obtainable, thereby providing a useful tool for various biomedical applications.
|
|
|
Channel and Nonchannel Forms of Spin-Labeled Gramicidin in Membranes and Their Equilibria
B.G. Dzikovski, P.P. Borbat, and J.H. Freed.
J. Phys. Chem. B 115, 176-185 (2011).
Supporting Information
<doi: 10.1021/jp108105k>
PMID:
21142163
PMCID:
PMC3076037
Publication #359
|

|
|
ABSTRACT: Channel and nonchannel forms of gramicidin A (GA) were studied by ESR in various lipid environments using new mono- and double-spin-labeled compounds. For GA channels, we demonstrate here how pulse dipolar ESR can be used to determine the orientation of the membrane-traversing molecule relative to the membrane normal and to study subtle effects of lipid environment on the interspin distance in the spin-labeled gramicidin channel. To study nonchannel forms of gramicidin, pulse dipolar ESR was used first to determine interspin distances corresponding to monomers and double-helical dimers of spin-labeled GA molecules in the organic solvents trifluoroethanol and octanol. The same distances were then observed in membranes. Since detection of nonchannel forms in the membrane is complicated by aggregation, we suppressed any dipolar spectra from intermolecular interspin distances arising from the aggregates by using double-labeled GA in a mixture with excess unlabeled GA. In hydrophobic mismatching lipids (Lβ phase of DPPC), gramicidin channels dissociate into free monomers. The backbone structure of the monomeric form is similar to a monomeric unit of the channel dimer. In addition to channels and monomers, the double-helical conformation of gramicidin is present in some membrane environments. In the gel phase of saturated phosphatidylcholines, the fraction of double helices increases in the following order: DLPC < DMPC < DSPC < DPPC. The equilibrium DHD/monomer ratio in DPPC was determined. In membranes, the double-helical form is present only in aggregates. In addition, we studied the effect of N-terminal substitution in the GA molecule upon channel formation. This work demonstrates how pulsed dipolar ESR may be utilized to study complex equilibria of peptides in membranes.
|
|
|
Stochastic Methods for Magnetic Resonance Spectroscopies
A. Polimeno, V. Barone, and J.H. Freed.
In Computational Strategies for Spectroscopy: From Small Molecules to Nano Systems; V. Barone, Ed.
Wiley: New York, NY, 2012; Chapter 12.
<doi: 10.1002/9781118008720.ch12>
PMID: [None - Book] PMCID:
[None - Book]
Publication #358
|

|
|
ABSTRACT: Physicochemical properties of molecules in solution depend on the action of different motions at several time and length scales, and information on multiscale dynamics can be gained, in principle, by a variety of spectroscopic techniques. In this work we review theoretical tools for the investigation of "slow" molecular motions, such as solvent cage effects in liquids and liquid crystals, global and local dynamics in proteins, reorientation dynamics, and internal (conformational) degrees of freedom. Spectroscopic techniques which are most sensitive to such motions are electron spin resonance and nuclear magnetic resonance and they require ad hoc theoretical treatment. In particular, we discuss the definition of multidimensional stochastic models and their treatment to interpret magnetic resonance spectroscopic data of rigid and flexible molecules in isotropic media, liquid crystals, and biosystems.
|
|
|
Comment on "The physical basis of model-free analysis of NMR relaxation data from proteins and complex fluids" [J. Chem. Phys. 131, 224507 (2009)]
E. Meirovitch, A. Polimeno, and J.H. Freed.
J. Chem. Phys. 132, 207101 (2010).
<doi: 10.1063/1.3429599>
PMID:
20515116
PMCID:
PMC2887917
Publication #357
|
|
|
In a recent article, Halle was critical of the use of the slowly relaxing local structure (SRLS) model for the analysis of NMR spin relaxation in proteins. Although he cited private communications with us, he did not discuss the contents of his article prior to its publication, nor were we aware of his intentions. We are in disagreement with a number of aspects of this article, but confine ourselves just to a brief rebuttal of his negative comments about SRLS and its use. We do, however, respect Halle’s efforts to provide a physical basis for the model-free (MF) approach.
|
|
|
A Multifrequency EPR Study of Fe2+ and Mn2+ Ions in a ZnSiF6·6H2O Single Crystal at Liquid-Helium Temperatures
S.K. Misra, S. Diehl, D. Tipikin, J.H. Freed.
J. Magn. Res. 205, 14-22 (2010).
<doi: 10.1016/j.jmr.2010.03.013>
PMID:
20395160
PMCID:
PMC2885525
Publication #356
|

|
|
ABSTRACT: A liquid-helium temperature study of Fe2+ and Mn2+ ions has been carried out on a single crystal of Fe2+-doped ZnSiF6⋅6H2O at 5–35 K at 170, 222.4 and 333.2 GHz. The spectra are found to be an overlap of two magnetically inequivalent Fe2+ ions, as well as that of an Mn2+ ion. From the simulation of the EPR line positions for the Fe2+ (d6, S = 2) ion the spin-Hamiltonian parameters were estimated for the two inequivalent Fe2+ ions at the various temperatures. From the relative intensities of lines the absolute sign of the fine-structure parameters have been estimated. In addition, the fine-structure and hyperfine-structure spin-Hamiltonian parameters for the Mn2+ ion, present as impurity at interstitial sites, were estimated from the hyperfine allowed and forbidden line positions. The particular virtues of such a single-crystal study vs. that on powders are noted.
|
|
|
Structure of the Ternary Complex Formed by a Chemotaxis Receptor Signaling Domain, the CheA Histidine Kinase, and the Coupling Protein CheW As Determined by Pulsed Dipolar ESR Spectroscopy
J. Bhatnagar, P.P. Borbat, A.M. Pollard, A.M. Bilwes, J.H. Freed, and B.R. Crane.
Biochemistry, 49, 3824-3841 (2010).
Supporting Information
<doi: 10.1021/bi100055m>
PMID:
20355710
PMCID:
PMC2873776
Publication #355
|

|
|
ABSTRACT: The signaling apparatus that controls bacterial chemotaxis is composed of a core complex containing chemoreceptors, the histidine autokinase CheA, and the coupling protein CheW. Site-specific spin labeling and pulsed dipolar ESR spectroscopy (PDS) have been applied to investigate the structure of a soluble ternary complex formed by Thermotoga maritima CheA (TmCheA), CheW, and receptor signaling domains. Thirty-five symmetric spin-label sites (SLSs) were engineered into the five domains of the CheA dimer and CheW to provide distance restraints within the CheA:CheW complex in the absence and presence of a soluble receptor that inhibits kinase activity (Tm14). Additional PDS restraints among spin-labeled CheA, CheW, and an engineered single-chain receptor labeled at six different sites allow docking of the receptor structure relative to the CheA:CheW complex. Disulfide cross-linking between selectively incorporated Cys residues finds two pairs of positions that provide further constraints within the ternary complex: one involving Tm14 and CheW and another involving Tm14 and CheA. The derived structure of the ternary complex indicates a primary site of interaction between CheW and Tm14 that agrees well with previous biochemical and genetic data for transmembrane chemoreceptors. The PDS distance distributions are most consistent with only one CheW directly engaging one dimeric Tm14. The CheA dimerization domain (P3) aligns roughly antiparallel to the receptor-conserved signaling tip but does not interact strongly with it. The angle of the receptor axis with respect to P3 and the CheW-binding P5 domains is bound by two limits differing by ˜20°. In one limit, Tm14 aligns roughly along P3 and may interact to some extent with the hinge region near the P3 hairpin loop. In the other limit, Tm14 tilts to interact with the P5 domain of the opposite subunit in an interface that mimics that observed with the P5 homologue CheW. The time domain ESR data can be simulated from the model only if orientational variability is introduced for the P5 and, especially, P3 domains. The Tm14 tip also binds beside one of the CheA kinase domains (P4); however, in both bound and unbound states, P4 samples a broad range of distributions that are only minimally affected by Tm14 binding. The CheA P1 domains that contain the substrate histidine are also broadly distributed in space under all conditions. In the context of the hexagonal lattice formed by trimeric transmembrane chemoreceptors, the PDS structure is best accommodated with the P3 domain in the center of a honeycomb edge.
|
|
|
Structural Dynamics of Bio-Macromolecules by NMR: The Slowly Relaxing Local Structure Approach
E. Meirovitch, Y.E. Shapiro, A. Polimeno, J.H. Freed.
Progress in NMR Spectroscopy, 56, 360-405 (2010).
<doi: 10.1016/j.pnmrs.2010.03.002>
PMID:
20625480
PMCID:
PMC2899824
Publication #354
|

|
|
INTRODUCTION: Protein dynamics by NMR has been reviewed extensively in recent years. These surveys show decisively that information on structure should be complemented by information on motion both to properly characterize the protein, and to understand its function. The time scale accessible by NMR extends from picoseconds to days, with different methods accessing different parts of this time axis. Here we focus on heteronuclear NMR spin relaxation used to study ps to ns protein dynamics. The slow limit of this time regime is determined by the global tumbling of the protein, with the rates for internal motion of the probe being typically faster.
|
|
|
Multifrequency Electron Spin Resonance Study of the Dynamics of Spin Labeled T4 Lysozyme
Z. Zhang, M.R. Fleissner, D.S. Tipikin, Z. Liang, J.K. Moscicki, K.A. Earle, W.L. Hubbell, and J.H. Freed.
J. Phys. Chem. B, 114, 5503-5521 (2010).
Supporting Information
<doi: 10.1021/jp910606h>
PMID:
20361789
PMCID:
PMC2865245
Publication #353
|

|
|
ABSTRACT: An extensive set of electron spin resonance spectra was obtained over a wide range of frequencies (9, 95, 170, and 240 GHz) and temperatures (2 to 32 °C) to explore the dynamic modes of nitroxide-labeled T4 lysozyme in solution. A commonly used nitroxide side chain (R1), or a methylated analogue with hindered internal motion (R2), was substituted for the native side chain at solvent-exposed helical sites, 72 or 131. The spectra at all four frequencies were simultaneously fit with the slowly relaxing local structure (SRLS) model. Good fits were achieved at all the temperatures. Two principle dynamic modes are included in the SRLS model, the global tumbling of the protein and the internal motion consisting of backbone fluctuations and side chain isomerizations. Three distinct spectral components were required for R1 and two for R2 to account for the spectra at all temperatures. One is a highly ordered and slow motional component, which is observed in the spectra of both R1 and R2; it may correspond to conformers stabilized by interaction with the protein surface. The fraction of this component decreases with increasing temperature and is more populated in the R2 spectra, possibly arising from stronger interaction of the nitroxide ring with the protein surface due to the additional methyl group. The other two components of R1 and the second component of R2 are characterized by fast anisotropic diffusion and relatively low ordering, most likely corresponding to conformers having little or no interactions with nearby residues. Ficoll of different concentrations was added to increase the solution viscosity, thereby slowing down the global tumbling of the protein. A significant effect of Ficoll on the internal motion of an immobilized component was apparent in R2 but not in R1. The ability of such multifrequency studies to separate the effects of faster internal modes of motion from slower overall motions is clearly demonstrated, and its utility in future studies is considered.
|
|
|
The Lipid-Binding Domain of Wild Type and Mutant α-Synuclein: Compactness and Interconversion Between the Broken and Extended Helix Forms
E.R. Georgieva, T.F. Ramlall, P.P. Borbat, J.H. Freed, and D. Eliezer.
J. Biol. Chem. 285, 28261-28274 (2010).
Supporting Information
<doi: 10.1074/jbc.M110.157214>
PMID:
20592036
PMCID:
PMC2934691
Publication #352
|

|
|
ABSTRACT: α-Synuclein (αS) is linked to Parkinson disease through its deposition in an amyloid fibril form within Lewy Body deposits, and by the existence of three αS point mutations that lead to early onset autosomal dominant Parkinsonism. The normal function of αS is thought to be linked to the ability of the protein to bind to the surface of synaptic vesicles. Upon binding to vesicles, αS undergoes a structural reorganization from a dynamic and disordered ensemble to a conformation consisting of a long extended helix. In the presence of small spheroidal detergent micelles, however, this extended helix conformation can convert into a broken helix state, in which a region near the middle of the helix unwinds to form a linker between the two resulting separated helices. Membrane-bound conformations of αS likely mediate the function of the protein, but may also play a role in the aggregation and toxicity of the protein. Here we have undertaken a study of the effects of the three known PD-linked mutations on the detergent- and membrane-bound conformations of αS, as well as factors that govern the transition of the protein between the extended helix and broken helix states. Using pulsed dipolar ESR measurements of distances up to 8.7 nm, we show that all three PD-linked αS mutants retain the ability to transition from the broken helix to the extended helix conformation. In addition, we find that the ratio of protein to detergent, rather than just the absolute detergent concentration, determines whether the protein adopts the broken or extended helix conformation.
|
|
|
Diphthamide Biosynthesis Requires an Organic Radical Generated by an Iron-Sulphur Enzyme
Y. Zhang, X. Zhu, A.T. Torelli, M. Lee, B. Dzikovski, R.M. Koralewski, E. Wang, J.H. Freed, C. Krebs, S.E. Ealick, and H. Lin.
Nature, 465, 891-896 (2010).
|

|
|
Diphthamide Biosynthesis Requires an Organic Radical Generated by an Iron-Sulphur Enzyme
Y. Zhang, X. Zhu, A.T. Torelli, M. Lee, B. Dzikovski, R.M. Koralewski, E. Wang, J.H. Freed, C. Krebs, S.E. Ealick, and H. Lin.
Nature, 465, 891-896 (2010).
Supporting Information
<doi: 10.1038/nature09138>
PMID:
20559380
PMCID:
PMC3006227
Publication #351
|

|
|
ABSTRACT: Archaeal and eukaryotic translation elongation factor 2 contain a unique post-translationally modified histidine residue called diphthamide, which is the target of diphtheria toxin. The biosynthesis of diphthamide was proposed to involve three steps, with the first being the formation of a C–C bond between the histidine residue and the 3-amino-3-carboxypropyl group of S-adenosyl-L-methionine (SAM). However, further details of the biosynthesis remain unknown. Here we present structural and biochemical evidence showing that the first step of diphthamide biosynthesis in the archaeon Pyrococcus horikoshii uses a novel iron–sulphur-cluster enzyme, Dph2. Dph2 is a homodimer and each of its monomers can bind a [4Fe–4S] cluster. Biochemical data suggest that unlike the enzymes in the radical SAM superfamily, Dph2 does not form the canonical 5′-deoxyadenosyl radical. Instead, it breaks the Cγ,Met–S bond of SAM and generates a 3-amino-3-carboxypropyl radical. Our results suggest that P. horikoshii Dph2 represents a previously unknown, SAM-dependent, [4Fe–4S]-containing enzyme that catalyses unprecedented chemistry.
|
|
|
Scanned-Probe Detection of Electron Spin Resonance from a Nitroxide Spin Probe
E.W. Moore, S.G. Lee, S.A. Hickman, S.J. Wright, L.E. Harrell, P.P. Borbat, J.H. Freed, and J.A. Marohn.
PNAS 106, 22251-22256 (2009).
Supporting Information
<doi: 10.1073/pnas.0908120106>
PMID:
20018707
PMCID:
PMC2799694
Publication #350
|
|
|
ABSTRACT: We report an approach that extends the applicability of ultrasensitive force-gradient detection of magnetic resonance to samples with spin-lattice relaxation times (T1) as short as a single cantilever period. To demonstrate the generality of the approach, which relies on detecting either cantilever frequency or phase, we used it to detect electron spin resonance from a T1 = 1 ms nitroxide spin probe in a thin film at 4.2 K and 0.6 T. By using a custom-fabricated cantilever with a 4 μm-diameter nickel tip, we achieve a magnetic resonance sensitivity of 400 Bohr magnetons in a 1 Hz bandwidth. A theory is presented that quantitatively predicts both the lineshape and the magnitude of the observed cantilever frequency shift as a function of field and cantilever-sample separation. Good agreement was found between nitroxide T1's measured mechanically and inductively, indicating that the cantilever magnet is not an appreciable source of spin-lattice relaxation here. We suggest that the new approach has a number of advantages that make it well suited to push magnetic resonance detection and imaging of nitroxide spin labels in an individual macromolecule to single-spin sensitivity.
|
|
|
Variable Coupling Scheme for High-Frequency Electron Spin Resonance Resonators Using Asymmetric Meshes
D.S. Tipikin, K.A. Earle and J.H. Freed.
Appl. Magn. Reson. 37, 819-832 (2010).
<doi: 10.1007/s00723-009-0088-1>
PMID:
20458356
PMCID:
PMC2865681
Publication #349
|
|
|
ABSTRACT: The sensitivity of a high-frequency electron spin resonance (ESR) spectrometer depends strongly on the structure used to couple the incident millimeter wave to the sample that generates the ESR signal. Subsequent coupling of the ESR signal to the detection arm of the spectrometer is also a crucial consideration for achieving high spectrometer sensitivity. In previous work, we found that a means for continuously varying the coupling was necessary for attaining high sensitivity reliably and reproducibly. We report here on a novel asymmetric mesh structure that achieves continuously variable coupling by rotating the mesh in its own plane about the millimeter-wave transmission-line optical axis. We quantify the performance of this device with nitroxide spin label spectra in both a lossy aqueous solution and a low-loss solid-state system. These two systems have very different coupling requirements and are representative of the range of coupling achievable with this technique. Lossy systems, in particular, are a demanding test of the achievable sensitivity and allow us to assess the suitability of this approach for applying high-frequency ESR, e.g., to the study of biological systems at physiological conditions. The variable coupling technique reported on here allows us to readily achieve a factor of ca. 7 improvement in the signal-to-noise ratio at 170 GHz and a factor of ca. 5 at 95 GHz over what has previously been reported for lossy samples.
|
|
|
Multifrequency ESR Study of Spin-Labeled Molecules in Inclusion Compounds with Cyclodextrins
B. Dzikovski, D. Tipikin, V. Livshits, K.A. Earle and J.H. Freed.
Phys. Chem. Chem. Phys. 11, 6676-6688 (2009).
Supporting Information
<doi: 10.1039/b903490k>
PMID:
19639141
PMCID:
PMC2743463
Publication #348
|

|
|
ABSTRACT: The molecular dynamics of spin-labeled compounds included into the solid phase of cyclodextrins (CDs) has been studied using conventional (X-band) ESR at 9 GHz and high-field high-frequency (HFHF) ESR at 240 and 170 GHz. The patterns of axial rotation at these higher frequencies are clear just by inspection of the spectrum, unlike the case for 9 GHz spectra. That is HFHF ESR is sensitive to molecular motion about the diffusion axis collinear with the X, Y or Z-direction of the magnetic g- and A-tensors of the nitroxide moiety (referred to, respectively, as X, Y or Z-rotation). For doxyl stearic acids (Z-rotation) and TEMPOyl caprylate (X-rotation) included in β- and γ-CDs we were able to determine the rate of molecular motion and the corresponding potential barriers. We emphasize that determining the rate of Z-rotation by ESR is feasible only using HFHF ESR. For the X-rotation case we suggest that the motion of the nitroxide moiety consists of fast small-angle librations about the magnetic X-axis superimposed by rotational diffusion about the same axis. The potential barrier of 1.7 Kcal mol−1 for this rotational diffusion is unusually low. A fascinating feature of TEMPO derivatives included in β-CD is the detectable molecular motion at temperatures below 77 K. For the other CD-spin probe systems, we used multifrequency analysis to assign the conformations of spin-labeled molecules. A dramatic spectral change for 16-sasl in β- and γ-CDs at ˜260 K corresponds to a tilting of the position of the nitroxide moiety on the rotating molecule relative to the long diffusion axis, while for TEMPO derivatives in γ-cyclodextrin below 200 K, we observe a rapid transition from fast to very slow rotational motion. More complex features are best studied by means of multifrequency ESR experiments. The visual clarity and the simplicity of analysis of the ESR spectra shown in this work should provide a benchmark for future studies of molecular motion by HFHF ESR.
|
|
|
Fusion Peptide from Influenza Hemagglutinin Increases Membrane Surface Order: An Electron-Spin Resonance Study
M. Ge and J.H. Freed.
Biophys. J. 96, 4925-4934 (2009).
Supporting Information
<doi: 10.1016/j.bpj.2009.04.015>
PMID:
19527651
PMCID:
PMC2712059
Publication #347
|

|
|
ABSTRACT: A spin-labeling study of interactions of a fusion peptide from the hemagglutinin of the influenza virus, wt20, and a fusion-inactive mutant ΔG1 with dimyristoylphosphatidylcholine (DMPC) and 1-palmitoyl-2-oleoyl-phosphatdylcholine bilayers was performed. We found that upon binding of wt20, the ordering of headgroups and the ordering of acyl chains near the headgroup increased significantly, in a manner consistent with a cooperative phenomenon. However, changes in the order at the end of the acyl chains were negligible. The ordering effect of wt20 on the headgroup was much stronger at pH 5 than at pH 7. No effect of ΔG1 binding on the order of bilayers was evident. We also found that 1-palmitoyl-2-hydroxyl phosphatidylcholine, a membrane-fusion inhibitor, decreased the ordering of DMPC headgroups, whereas arachidonic acid, a membrane-fusion promoter, increased the ordering of DMPC headgroups. These results suggest that increases in headgroup ordering may be important for membrane fusion. We propose that upon binding of wt20, which is known to affect only the outer leaflet of the bilayer, this outer leaflet becomes more ordered, and thus more solid-like. Then the coupling between the hardened outer leaflet and the softer inner leaflet generates bending stresses in the bilayer, which tend to increase the negative curvature of the bilayer. We suggest that the increased ordering in the headgroup region enhances dipolar interactions and lowers electrostatic energy, which may provide an energy source for membrane fusion. Possible roles of bending stresses in promoting membrane fusion are discussed.
|
|
|
Molecular-Scale Force Measurement in a Coiled-Coil Peptide Dimer by Electron Spin Resonance
S.V. Gullà, G. Sharma, P.Borbat, J.H. Freed, H. Ghimire, M.R. Benedikt, N.L. Holt, G.A. Lorigan, K. Rege, C. Mavroidis, and D.E. Budil.
J. Am. Chem. Soc. 131, 5374-5375 (2009).
|

|
|
Molecular-Scale Force Measurement in a Coiled-Coil Peptide Dimer by Electron Spin Resonance
S.V. Gullà, G. Sharma, P.Borbat, J.H. Freed, H. Ghimire, M.R. Benedikt, N.L. Holt, G.A. Lorigan, K. Rege, C. Mavroidis, and D.E. Budil.
J. Am. Chem. Soc. 131, 5374-5375 (2009).
Supporting Information
<doi: 10.1021/ja900230w>
PMID:
19331323
PMCID:
PMC2888838
Publication #346
|

|
|
ABSTRACT: A new method for measuring forces between small protein domains based on double electron−electron resonance (DEER) spectroscopy is demonstrated using a model peptide derived from the α-helical coiled-coil leucine zipper of yeast transcriptional activator GCN4. The equilibrium distribution of distances between two nitroxide spin labels rigidly attached to the helices of the dimer was determined by DEER and yielded a closing force of 100 ± 10 pN between monomers, in excellent agreement with theoretical predictions.
|
|
|
Calculation of Double-Quantum-Coherence Two-dimensional Spectra: Distance Measurements and Orientational Correlations
S.K. Misra, P.P. Borbat, J.H. Freed.
Appl. Magn. Reson. 36, 237-258 (2009).
<doi: 10.1007/s00723-009-0023-5>
PMID:
20161423
PMCID:
PMC2786184
Publication #345
|

|
|
ABSTRACT: The double-quantum-coherence (DQC) echo signal for two coupled nitroxides separated by distances ≳10 Å, is calculated rigorously for the six-pulse sequence. Successive application of six pulses on the initial density matrix, with appropriate inter-pulse time evolution and coherence pathway selection leaves only the coherent pathways of interest. The amplitude of the echo signal following the last π pulse can be used to obtain a one-dimensional (1D) dipolar spectrum (Pake doublet), and the echo envelope can be used to construct the 2D DQC spectrum. The calculations are carried out using the product space spanned by the two electron-spin magnetic quantum numbers m1, m2 and the two nuclear-spin magnetic quantum numbers M1, M2, describing, e.g. two coupled nitroxides in bilabeled proteins. The density matrix is subjected to a cascade of unitary transformations taking into account dipolar and electron exchange interactions during each pulse and during the evolution in the absence of a pulse. The unitary transformations use the eigensystem of the effective spin Hamiltonians obtained by numerical matrix diagonalization. Simulations are carried out for a range of dipolar interactions, D, and microwave magnetic field strength B1 for both fixed and random orientations of the two 14N (and 15N) nitroxides. Relaxation effects were not included. Several examples of 1D and 2D Fourier transforms of the time-domain signals versus dipolar evolution and spin-echo envelope time variables are shown for illustration. Comparisons are made between 1D rigorous simulations and analytical approximations. The rigorous simulations presented here provide insights into DQC electron spin resonance spectroscopy, they serve as a standard to evaluate the results of approximate theories, and they can be employed to plan future DQC experiments.
|
|
|
A Scissors Mechanism for Stimulation of SNARE-Mediated Lipid Mixing by Cholesterol
J. Tong, P.P. Borbat, J.H. Freed, and Y.-K. Shin.
PNAS 106, 5141-5146 (2009).
Supporting Information
<doi: 10.1073/pnas.0813138106>
PMID:
19251653
PMCID:
PMC2663986
Publication #344
|
|
|
ABSTRACT: Neurotransmitter release at the synapse requires membrane fusion. The SNARE complex, composed of the plasma membrane t-SNAREs syntaxin 1A and SNAP-25 and the vesicle v-SNARE synaptobrevin, mediates the fusion of 2 membranes. Synaptic vesicles contain unusually high cholesterol, but the exact role of cholesterol in fusion is not known. In this study, cholesterol was found to stimulate SNARE-mediated lipid mixing of proteoliposomes by a factor of 5 at a physiological concentration. Surprisingly, however, the stimulatory effect was more pronounced when cholesterol was on the v-SNARE side than when it was on the t-SNARE side. Site-directed spin labeling and both continuous wave (CW) and pulsed EPR revealed that cholesterol induces a conformational change of the v-SNARE transmembrane domain (TMD) from an open scissors-like dimer to a parallel dimer. When the TMD was forced to form a parallel dimer by the disulfide bond, the rate was stimulated 2.3-fold even without cholesterol, supporting the relevance of the open-to-closed conformational change to the fusion activity. The open scissors-like conformation may be unfavorable for fusion and cholesterol may relieve this inhibitory factor.
|
|
|
Determination of Tie-Line Fields for Coexisting Lipid Phases: An ESR Study
A.K. Smith and J.H. Freed.
J. Phys. Chem. B 113, 3957-3971 (2009).
<doi: 10.1021/jp808412x>
PMID:
19673072
PMCID:
PMC2772187
Publication #343
|

|
|
ABSTRACT: A novel method we refer to as the tie-line field (TLF) method has been developed to globally determine the tie lines of any three-component two-phase coexistence region by fitting electron-spin resonance (ESR) spectra obtained from compositions on the coexistence curve and within the coexistence region. The TLF method is illustrated by applying it to the liquid-ordered (Lo) and liquid-disordered (Ld) phase coexistence region of the lipid system brain-sphingomyelin / dioleoylphosphatidylcholine / cholesterol (SPM/DOPC/chol), for which an estimate of a tie-line was previously obtained by an earlier method also using ESR spectra. The essential aspect of the TLF method is the unique parametrization of the coexistence region called a "ruled surface". The use of the ruled surface enables one to guarantee that tie lines do not cross, as required by the phase rule, whereas previous methods lack this important constraint. It also makes simultaneous use of the full data set in determining the TLF and leads to a more efficient experimental design than previously used. The method is first tested out on synthetic data sets, then least-squares fitting of the ESR spectra with the parametrized model results in a tie-line field consistent with other known information on this lipid system. The best-fit tie-line field consists of the set of tie lines which are not exactly parallel; they exhibit a gradual change in slope with the largest slope within the coexistence region connecting the coexistence curve compositions with the highest and lowest cholesterol concentrations. The results are compared with those from more constrained methods of representing the tie-line fields as well as with the previous tie-line determination for the SPM/DOPC/chol system. An accurate determination of the tie-line field of phase coexistence regions in lipid systems is a necessary step in determining coexisting lipid compositions to serve as models of cell plasma membranes.
|
|
|
Multifrequency Electron Spin Resonance Spectra of a Spin-Labeled Protein Calculated from Molecular Dynamics Simulations
D. Sezer, J.H. Freed, and B. Roux.
J. Am. Chem. Soc. 131, 2597-2605 (2009).
Supporting Information
<doi: 10.1021/ja8073819>
PMID:
19191603
PMCID:
PMC2763593
Publication #342
|

|
|
ABSTRACT: Multifrequency electron spin resonance (ESR) spectra provide a wealth of structural and dynamic information about the local environment of the spin label and, indirectly, about the spin-labeled protein. Relating the features of the observed spectra to the underlying molecular motions and interactions is, however, challenging. To make progress toward a rigorous interpretation of ESR spectra, we perform extensive molecular dynamics (MD) simulations of fully solvated T4 Lysozyme, labeled with the spin label MTSSL at positions 72 and 131. These two sites have been the object of numerous experimental studies and are generally considered as prototypical solvent-exposed sites on the surfaces of α-helices. To extend the time window afforded by the MD simulations, stochastic Markov models reflecting the dynamics of the spin label side chains in terms of their rotameric states are constructed from the trajectories. The calculated multifrequency ESR spectra are in very good agreement with experiment for three different magnetic field strengths without adjusting any parameters. During the trajectories, the spin labels interconvert among a fairly large number of conformations and display a propensity to form interactions with protein residues other than their nearest neighbors along the helix. The detailed picture of the spin label emerging from the MD simulations provides useful insight into the molecular origins of the available spectroscopic and crystallographic data.
|
|
|
A 236 GHz Fe3+ EPR Study of Nanoparticles of the Ferromagnetic Room-Temperature Semiconductor Sn1-xFexO2 (x = 0.005)
S.K. Misra, S.I. Andronenko, A. Punnoose, D. Tipikin, and J.H. Freed.
Appl. Magn. Reson. 36, 291-295 (2009).
<doi: 10.1007/s00723-009-0024-4>
PMID:
20161547
PMCID:
PMC2805097
Publication #341
|

|
|
ABSTRACT: High-frequency (236 GHz) electron paramagnetic resonance (EPR) studies of Fe3+ ions at 255 K are reported in a Sn1−xFexO2 powder with x = 0.005, which is a ferromagnetic semiconductor at room temperature. The observed EPR spectrum can be simulated reasonably well as the overlap of spectra due to four magnetically inequivalent high-spin (HS) Fe3+ ions (S = 5/2). The spectrum intensity is calculated, using the overlap I(BL) + (I(HS1) + I(HS2) + I(HS3) + I(HS4)) × exp(−0.00001B), where B is the magnetic field intensity in Gauss, I represents the intensity of an EPR line (HS1, HS2, HS3, HS4), and BL stands for the baseline (the exponential factor, as found by fitting to the experimental spectrum, is related to the Boltzmann population distribution of energy levels at 255 K, which is the temperature of the sample in the spectrometer). These high-frequency EPR results are significantly different from those at X-band. The large values of the zero-field splitting parameter (D) observed here for the four centers at the high frequency of 236 GHz are beyond the capability of X-band, which can only record spectra of ions with much smaller D values than those reported here.
|
|
|
Membrane-Bound α-Synuclein Forms an Extended Helix: Long-Distance Pulsed ESR Measurements Using Vesicles, Bicelles, and Rodlike Micelles
E.R. Georgieva, T.F. Ramlall, P.P. Borbat, J.H. Freed, and D. Eliezer.
J. Am. Chem. Soc. 130, 12856-12857 (2008).
Supporting Information
<doi: 10.1021/ja804517m>
PMID:
18774805
PMCID:
PMC2626176
Publication #340
|
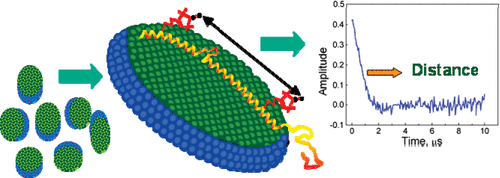
|
|
ABSTRACT: We apply pulsed dipolar ESR spectroscopy (Ku-band DEER) to elucidate the global conformation of the Parkinson's disease-associated protein, α-synuclein (αS) bound to small unilamellar phospholipid vesicles, rodlike SDS micelles, or lipid bicelles. By measuring distances as long as ˜7 nm between introduced pairs of nitroxide spin labels, we show that distances are close to the expectations for a single continuous helix in all cases studied. In particular, we find distances of 7.5 nm between sites 24 and 72; 5.5 nm between sites 24 and 61; and 2 nm between sites 35 and 50. We conclude that αS does not retain a "hairpin" structure with two antiparallel helices, as is known to occur with spheroidal micelles, in agreement with our earlier finding that the protein's geometry is determined by the surface topology rather than being constrained by the interhelix linker. While the possibility of local helix discontinuities in the structure of membrane-bound αS remains, our data are more consistent with one intact helix. Importantly, we demonstrate that bicelles produce very similar results to liposomes, while offering a major improvement in experimentally accessible distance range and resolution, and thus are an excellent lipid membrane mimetic for the purpose of pulse dipolar ESR spectroscopy.
|
|
|
Using Markov Models to Simulate Electron Spin Resonance Spectra from Molecular Dynamics Trajectories
D. Sezer, J.H. Freed, and B. Roux.
J. Phys. Chem. B 112, 11014-11027 (2008).
<doi: 10.1021/jp801608v>
PMID:
18698714
PMCID:
PMC2562300
Publication #339
|
|
|
ABSTRACT: Simulating electron spin resonance (ESR) spectra directly from molecular dynamics simulations of a spin-labeled protein necessitates a large number (hundreds or thousands) of relatively long (hundreds of nanoseconds) trajectories. To meet this challenge, we explore the possibility of constructing accurate stochastic models of the spin label dynamics from atomistic trajectories. A systematic, two-step procedure, based on the probabilistic framework of hidden Markov models, is developed to build a discrete-time Markov chain process that faithfully captures the internal spin label dynamics on time scales longer than about 150 ps. The constructed Markov model is used both to gain insight into the long-lived conformations of the spin label and to generate the stochastic trajectories required for the simulation of ESR spectra. The methodology is illustrated with an application to the case of a spin-labeled poly alanine α helix in explicit solvent.
|
|
|
Parametrization, Molecular Dynamics Simulation, and Calculation of Electron Spin Resonance Spectra of a Nitroxide Spin Label on a Polyalanine α-Helix
D. Sezer, J.H. Freed, B. Roux.
J. Phys. Chem. B 112, 5755-5767 (2008).
Supporting Information
<doi: 10.1021/jp711375x>
PMID:
18412413
PMCID:
PMC2766176
Publication #338
|

|
|
ABSTRACT: The nitroxide spin label 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl-methanethiosulfonate (MTSSL), commonly used in site-directed spin labeling of proteins, is studied with molecular dynamics (MD) simulations. After developing force field parameters for the nitroxide moiety and the spin label linker, we simulate MTSSL attached to a polyalanine α-helix in explicit solvent to elucidate the factors affecting its conformational dynamics. Electron spin resonance spectra at 9 and 250 GHz are simulated in the time domain using the MD trajectories and including global rotational diffusion appropriate for the tumbling of T4 Lysozyme in solution. Analysis of the MD simulations reveals the presence of significant hydrophobic interactions of the spin label with the alanine side chains.
|
|
|
Simulating Electron Spin Resonance Spectra of Nitroxide Spin Labels from Molecular Dynamics and Stochastic Trajectories
D. Sezer, J.H. Freed, and B. Roux.
J. Chem. Phys. 128, 165106, (2008).
<doi: 10.1063/1.2908075>
PMID:
18447510
PMCID:
PMC2809671
Publication #337
|

|
|
ABSTRACT: Simulating electron spin resonance spectra of nitroxide spin labels from motional models is necessary for the quantitative analysis of experimental spectra. We present a framework for modeling the spin label dynamics by using trajectories such as those from molecular dynamics (MD) simulations combined with stochastic treatment of the global protein tumbling. This is achieved in the time domain after two efficient numerical integrators are developed: One for the quantal dynamics of the spins and the other for the classical rotational diffusion. For the quantal dynamics, we propagate the relevant part of the spin density matrix in Hilbert space. For the diffusional tumbling, we work with quaternions, which enables the treatment of anisotropic diffusion in a potential expanded as a sum of spherical harmonics. Time-averaging arguments are invoked to bridge the gap between the smaller time step of the MD trajectories and the larger time steps appropriate for the rotational diffusion and/or quantal spin dynamics.
|
|
|
Membrane Fluidity
B. Dzikovski and J.H. Freed.
In Wiley Encyclopedia of Chemical Biology; T. P. Begley and B. A. Baird, Eds.
Wiley; Hoboken, NJ, USA, 2009; Chapter 2, pp 728-741.
|
|
|
Membrane Fluidity
B. Dzikovski and J.H. Freed.
In Wiley Encyclopedia of Chemical Biology; T. P. Begley and B. A. Baird, Eds.
Wiley; Hoboken, NJ, USA, 2009; Chapter 2, pp 728-741.
<doi: 10.1002/9780470048672.wecb313>
PMID: [None - Book] PMCID:
[None - Book]
Publication #336
|

|
|
ABSTRACT: In the popular fluid mosaic model for biomembranes, membrane proteins and other membrane-embedded molecules are in a two-dimensional fluid formed by the phospholipids. Such a fluid state allows free motion of constituents within the membrane bilayer and is extremely important for membrane function. The term "membrane fluidity" is a general concept, which refers to the ease of motion for molecules in the highly anisotropic membrane environment. We give a brief description of physical parameters associated with membrane fluidity, such as rotational and translational diffusion rates, order parameters, etc., and review physical methods used for their determination. We also show limitations of the fluid mosaic model and discuss recent developments in membrane science that pertain to fluidity, such as evidence for compartmentalization of the biomembrane by the cell cytoskeleton.
|
|
|
Determination of the Oligomeric States of Human and Rat Monoamine Oxidases in the Outer Mitochondrial Membrane and Octyl β-D-Glucopyranoside Micelles Using Pulsed Dipolar Electron Spin Resonance Spectroscopy
A.K. Upadhyay, P.P. Borbat, J. Wang, J.H. Freed, and D.E. Edmondson.
Biochemistry 47, 1554-1566 (2008).
<doi: 10.1021/bi7021377>
PMID:
18198902
PMCID:
[accepted before 04/08]
Publication #335
|

|
|
ABSTRACT: Human monoamine oxidase A (hMAOA) is considered to be unique among mammalian MAOs in having a non-conservative Glu-X-Lys mutation (X being 151 in MAOAs and 142 in MAOB's), which is suggested to be the reason for its monomeric structure. This hypothesis has been tested in this work. A pargyline based nitroxide spin labeled irreversible inhibitor (ParSL) was used as a MAO active site specific spin probe to measure intersubunit distances in detergent (octyl β-D-glucopyranoside, OGP) purified and OMM bound forms by a pulsed dipolar ESR spectroscopic (PDS) technique. In a parallel approach, the covalent flavin cofactor present in the MAO active sites was reduced to its respective anionic flavin semiquinone and used for measuring inter-flavin distances in detergent purified samples. The measured interspin distances are within 0.1–0.3 nm of those estimated from the available dimeric crystal structures of human MAOB and rat MAOA and show that all human and rat MAOs exist as dimers in the OMM. In the OGP micelle, however, human and rat MAOAs exist only partially (≤50%) as dimers, whereas human and rat MAOBs exist entirely as dimers. The Lys-151-Glu mutant of human MAOA and the Glu-142-Lys mutant of human MAOB exhibit similar spectral properties as the corresponding wild-type enzymes. Therefore, no role of the Glu-X residue in stabilizing dimeric structures of MAOs was found. The monomeric crystal structure reported for human MAOA is thus a result of its instability in the OGP micelles. The general applicability of the PDS technique to structural studies of membrane proteins in their native membrane environments and detergent purified forms is discussed.
|
|
|
Pros and Cons of Pulse Dipolar ESR: DQC and DEER
P.P. Borbat and J.H. Freed.
EPR Newsletter 17, 21-33 (2007).
Publication #334
|

|
|
INTRODUCTION: Continuous-wave (cw) and pulsed ESR have been extensively applied to biological problems in the context of molecular dynamics and are now increasingly applied to study biomolecular structure and function. cw ESR has been used to measure distances in the range of 6–20 Å between pairs of nitroxide spin labels. Distance measurements using pulse ESR methods, a major advance in this area, are currently able to deliver long-distance constraints in the range of 10–80 Å. The distance constraints from pulse ESR can for example be used to establish protein folding or orient and dock proteins and their subunits, yielding useful insights into the structure of a protein or a protein complex. They can also aid in refinement of NMR data. We refer to this emerging methodology as 'pulsed dipolar (ESR) spectroscopy' or PDS for short.
|
|
|
An Improved Picture of Methyl Dynamics in Proteins from Slowly Relaxing Local Structure Analysis of 2H Spin Relaxation
E. Meirovitch, Y.E. Shapiro, A. Polimeno, and J.H. Freed.
J. Phys. Chem. B 111, 12865-12875 (2007).
<doi: 10.1021/jp072156s>
PMID:
17941658
PMCID:
PMC2885794
Publication #333
|

|
|
ABSTRACT: Protein dynamics is intimately related to biological function. Core dynamics is usually studied with 2H spin relaxation of the 13CDH2 group, analyzed traditionally with the model-free (MF) approach. We showed recently that MF is oversimplified in several respects. This includes the assumption that the local motion of the dynamic probe and the global motion of the protein are decoupled, the local geometry is simple, and the local ordering is axially symmetric. Because of these simplifications MF has yielded a puzzling picture where the methyl rotation axis is moving rapidly with amplitudes ranging from nearly complete disorder to nearly complete order in tightly packed protein cores. Our conclusions emerged from applying to methyl dynamics in proteins the slowly relaxing local structure (SRLS) approach of Polimeno and Freed (Polimeno, A.; Freed, J. H. J. Phys. Chem. 1995, 99, 10995–11006.), which can be considered the generalization of MF, with all the simplifications mentioned above removed. The SRLS picture derived here for the B1 immunoglobulin binding domain of peptostreptococcal protein L, studied over the temperature range of 15–45 °C, is fundamentally different from the MF picture. Thus, methyl dynamics is characterized structurally by rhombic local potentials with varying symmetries and dynamically by tenfold slower rates of local motion. On average, potential rhombicity decreases, mode-coupling increases, and the rate of local motion increases with increasing temperature. The average activation energy for local motion is 2.0 ± 0.2 kcal/mol. Mode-coupling affects the analysis even at 15 °C. The accuracy of the results is improved by including in the experimental data set relaxation rates associated with rank 2 coherences.
|
|
|
Conformational Motion of the ABC Transporter MsbA Induced by ATP Hydrolysis
P.P. Borbat, K. Surendhran, M. Bortolus, P. Zou, J.H. Freed, H.S. Mchaourab.
PLoS Biology 5, 2211-2219 (2007).
Supporting Information
<doi: 10.1371/journal.pbio.0050271>
PMID:
17927448
PMCID:
PMC2001213
Publication #332
|

|
|
ABSTRACT: We measured the amplitude of conformational motion in the ATP-binding cassette (ABC) transporter MsbA upon lipopolysaccharide (LPS) binding and following ATP turnover by pulse double electron-electron resonance and fluorescence homotransfer. The distance constraints from both methods reveal large-scale movement of opposite signs in the periplasmic and cytoplasmic part of the transporter upon ATP hydrolysis. LPS induces distinct structural changes that are inhibited by trapping of the transporter in an ATP post-hydrolysis intermediate. The formation of this intermediate involves a 33-Å distance change between the two ABCs, which is consistent with a dimerization-dissociation cycle during transport that leads to their substantial separation in the absence of nucleotides. Our results suggest that ATP-powered transport entails LPS sequestering into the open cytoplasmic chamber prior to its translocation by alternating access of the chamber, made possible by 10–20-Å conformational changes.
|
|
|
Dynamic Molecular Structure and Phase Diagram of DPPC-Cholesterol Binary Mixtures: A 2D-ELDOR Study
Y.-W. Chiang, A.J. Costa-Filho, and J.H. Freed.
J. Phys. Chem. B 111, 11260-11270 (2007).
Supporting Information
<doi: 10.1021/jp0732110>
PMID:
17760438
Publication #331
|

|
|
ABSTRACT: This paper is an application of 2D electron−electron double resonance (2D-ELDOR) with the "full Sc− method" to study model membranes. We obtain and confirm the phase diagram of 1,2-dipalmitoyl-sn-glycerophosphatidylcholine (DPPC)−cholesterol binary mixtures versus temperature and provide quantitative descriptions for its dynamic molecular structure using 2D-ELDOR at the Ku band. The spectra from the end-chain 16-PC spin label in multilamellar phospholipid vesicles are obtained for cholesterol molar concentrations ranging from 0 to 50% and from 25 to 60 °C. This phase diagram consists of liquid-ordered, liquid-disordered, and gel phases and phase coexistence regions. The phase diagram is carefully examined according to the spectroscopic evidence, and the rigorous interpretation for the line shape changes. We show that the 2D-ELDOR spectra differ markedly with variation in the composition. The extensive line shape changes in the 2D-plus-mixing-time representation provide useful information to define and characterize the membrane phases with respect to their dynamic molecular structures and to determine the phase boundaries. The homogeneous T2's are extracted from the pure absorption spectra and are used to further distinguish the membrane phases. These results show 2D-ELDOR to be naturally suitable for probing and reporting the dynamic structures of microdomains in model membrane systems and, moreover, providing a very detailed picture of their molecular dynamic structure, especially with the aid of the "full Sc− method".
|
|
|
Rigid Body Refinement of Protein Complexes with Long-Range Distance Restraints from Pulsed Dipolar ESR
J. Bhatnagar, J.H. Freed, and B.R. Crane.
In Two-Component Signaling Systems, Part B, M. Simon, B.R. Crane and A.B. Crane, Eds.
Methods in Enzymology 423, Ch. 4, pp. 117-133 (2007).
<doi: 10.1016/S0076-6879(07)23004-6>
PMID:
17609128
Publication #330
|

|
|
ABSTRACT: The modeling of protein–protein complexes greatly benefits from the incorporation of experimental distance restraints. Pulsed dipolar electron spin resonance spectroscopy is one such powerful technique for obtaining long-distance restraints in protein complexes. Measurements of the dipolar interaction between two spins placed specifically within a protein complex give information about the spin–spin separation distance. We have developed a convenient method to incorporate such long-range distance information in the modeling of protein–protein complexes that is based on rigid body refinement of the protein components with the software Crystallography and NMR System (CNS). Factors affecting convergence such as number of restraints, error allocation scheme, and number and position of spin labeling sites were investigated with real and simulated data. The use of 4 to 5 different labeling sites on each protein component was found to provide sufficient coverage for producing accuracies limited by the uncertainty in the spin-label conformation within the complex. With an asymmetric scheme of allocating this uncertainty, addition of simulated restraints revealed the importance of longer distances within a limited set of total restraints. We present two case studies: (1) refinement of the complex formed between the histidine kinase CheA and its coupling protein CheW, and (2) refinement of intra-helical separations in the protein a-synuclein bound to micelles.
|
|
|
Measuring Distances by Pulsed Dipolar ESR Spectroscopy: Spin-Labeled Histidine Kinases
P.P. Borbat and J.H. Freed.
In Two-Component Signaling Systems, Part B, M. Simon, B.R. Crane and A.B. Crane, Eds.
Methods in Enzymology 423, Ch. 3, pp. 52-116 (2007).
<doi: 10.1016/S0076-6879(07)23003-4>
PMID:
17609127
Publication #329
|
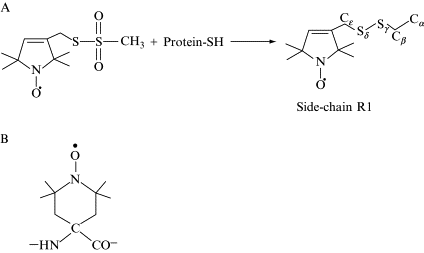
|
|
ABSTRACT: Applications of dipolar ESR spectroscopy to structural biology are rapidly expanding, and it has become a useful method that is aimed at resolving protein structure and functional mechanisms. The method of pulsed dipolar ESR spectroscopy (PDS) is outlined in the first half of the chapter, and it illustrates the simplicity and potential of this developing technology with applications to various biological systems. A more detailed description is presented of the implementation of PDS to reconstruct the ternary structure of a large dimeric protein complex from Thermotoga maritima, formed by the histidine kinase CheA and the coupling protein CheW. This protein complex is a building block of an extensive array composed of coupled supramolecular structures assembled from CheA/CheW proteins and transmembrane signaling chemoreceptors, which make up a sensor that is key to controlling the motility in bacterial chemotaxis. The reconstruction of the CheA/CheW complex has employed several techniques, including X-ray crystallography and pulsed ESR. Emphasis is on the role of PDS, which is part of a larger effort to reconstruct the entire signaling complex, including chemoreceptor, by means of PDS structural mapping. In order to precisely establish the mode of coupling of CheA to CheW and to globally map the complex, approximately 70 distances have already been determined and processed into molecular coordinates by readily available methods of distance geometry constraints.
|
|
|
2D-ELDOR Using Full Sc- Fitting and Absorption Lineshapes
Y.-W. Chiang, A. Costa-Filho, J.H. Freed.
J. Magn. Reson. 188, 231-245 (2007).
<doi: 10.1016/j.jmr.2007.06.014>
PMID:
17681478
Publication #328
|

|
|
ABSTRACT: Recent progress in developing 2D-ELDOR (2D electron–electron double resonance) techniques to better capture molecular dynamics in complex fluids, particularly in model and biological membranes, is reported. The new "full Sc− method", which corrects the spectral analysis for the phase distortion effects present in the experiments, is demonstrated to enhance the sensitivity of 2D-ELDOR in reporting on molecular dynamics in complex membrane environments. That is, instead of performing spectral fitting in the magnitude mode, our new method enables simultaneous fitting of both the real and imaginary components of the Sc− signal. The full Sc− fitting not only corrects the phase distortions in the experimental data but also more accurately determines instrumental dead times. The phase corrections applied to the Sc− spectrum enable the extraction of the pure absorption-mode spectrum, which is characterized by much better resolution than the magnitude-mode spectrum. In the absorption mode, the variation of homogeneous broadening, which reports on the dynamics of the spin probe, can even be observed by visual inspection. This new method is illustrated with results from model membranes of dipalmitoyl-sn-glycero-phosphatidylcholine (DPPC)–cholesterol binary mixtures, as well as with results from plasma membrane vesicles of mast cells. In addition to the dynamic parameters, which provide quantitative descriptions for membranes at the molecular level, the high-resolution absorption spectra themselves may be used as a "fingerprint" to characterize membrane phases and distinguish coexisting components in biomembranes. Thus we find that 2D-ELDOR is greatly improved with the new "full Sc− method" especially for exploring the complexity of model and biological membranes.
|
|
|
Two-Dimensional ELDOR in the Study of Model and Biological Membranes
Y.-W. Chiang, A.J. Costa-Filho, and J.H. Freed.
Appl. Magn. Reson. 31, 375-386 (2007).
<doi: 10.1007/BF03166591>
Publication #327
|

|
|
ABSTRACT: Recent studies on model and biological membranes by two-dimensional (2-D) electronelectron double resonance (ELDOR) are reviewed and discussed. The studies include (1) the phase behavior of dispersions of phospholipid-cholesterol membrane vesicles; (2) the effect of the ion-channel-forming peptide gramicidin A on the lipid membrane; and (3) the effects of stimulation by antigen of the immunoglobulin E receptors in plasma membrane vesicles upon the lipid structure. In the first studies it is shown that the 2-D ELDOR spectra enable clear distinctions amongst the different phases, and this leads to a reliable temperature and composition-dependent phase diagram. In the second studies it is possible to distinguish bulk and boundary lipids and to describe their different dynamic structures. In the third studies we could distinguish both liquid-ordered and liquid-disordered spectral components, and one finds that the fraction of the latter increases as a result of stimulation. Emphasis is placed on the new "full Sc — method" of processing the 2-D ELDOR data to significantly enhance spectral resolution, which is particularly important in studying the spectra from coexisting phases or components. In the full Sc — method, one utilizes both the real and imaginary parts of the signal, instead of their magnitude; the needed phase corrections are obtained as part of the nonlinear least-squares fitting of the 2-D ELDOR data.
|
|
|
|
|
Collective Fluctuations in Ordered Fluids Investigated by Two-Dimensional Electron-Electron Double Resonance Spectroscopy
B. Fresch, D. Frezzato, G.J. Moro, G. Kothe, and J.H. Freed.
J. Phys. Chem. B 110, 24238-24254, (2006).
<doi: 10.1021/jp064028u>
PMID:
17125397
Publication #325
|
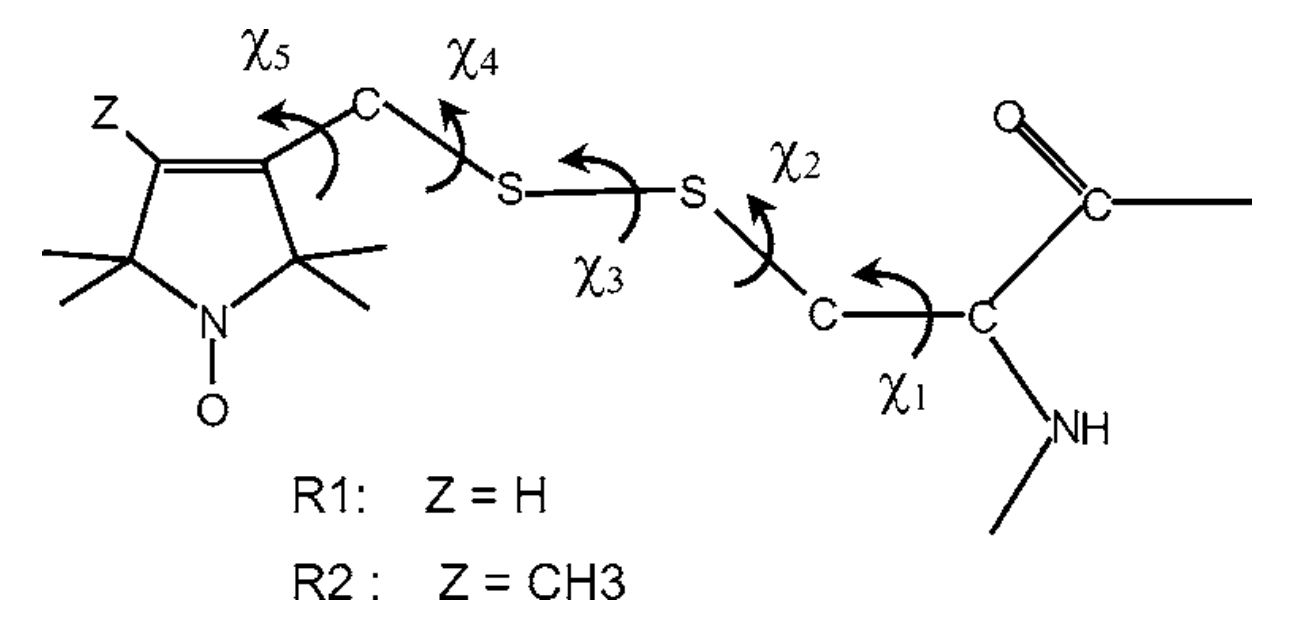
|
|
ABSTRACT: Two-dimensional electron−electron double resonance (2D-ELDOR) is a technique that is sensitive to the dynamical processes affecting spin labels in complex fluid environments. In ordered fluids, such as membrane vesicles, the 2D-ELDOR experiment is affected by the molecular tumbling in the locally ordered environment. This motion occurs on two different time scales, the faster molecular motion relative to the local director, and the slower collective fluctuations of the director field. In the experimental study of Patyal, Crepeau, and Freed (Biophys. J. 1997, 73, 2201), it was found that the widths of the autopeaks of the 2D-ELDOR spectrum increased as a function of the mixing time. In the present work, a theory is developed for the effects of director fluctuations on the autopeaks in the 2D-ELDOR experiment by employing an analytical solution of the stochastic Liouville equation for which the director field is treated as a multidimensional Gaussian process, as previously developed by Frezzato, Kothe, and Moro (J. Phys. Chem. B 2001, 105, 1281 and J. Phys. Chem. B 2004, 108, 9505). Good agreement is found between theory and experiment, notably the only adjustable parameter is κ, the bending elastic modulus of the membrane. The values of κ = 11 × 10-20 J for 1,2-dipalmitoyl-sn-glycero-phosphatidylcholine (DPPC) vesicles and κ = 15 × 10-20 J for DPPC/gramicidin A (5:1) vesicles, both at 45 °C, were found from the analysis and agree well with previous related measurements by other physical techniques. This establishes 2D-ELDOR as a useful technique to study the elastic properties of biological membranes.
|
|
|
Modeling the Effects of Structure and Dynamics of the Nitroxide Side Chain on the ESR Spectra of Spin-Labeled Proteins
F. Tombolato, A. Ferrarini, and J.H. Freed.
J. Phys. Chem. B 110, 26260-26271 (2006).
<doi: 10.1021/jp064028u>
PMID:
17181284
PMCID:
PMC2885803
Publication #324
|
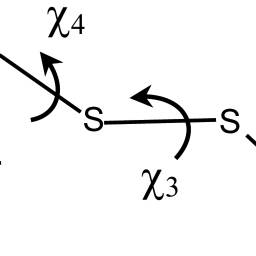
|
|
ABSTRACT: In the companion paper (J. Phys. Chem. B 2006, 110, jp0629487), a study of the conformational dynamics of methanethiosulfonate spin probes linked at a surface-exposed α-helix has been presented. Here, on the basis of this analysis, X-band ESR spectra of these spin labels are simulated within the framework of the Stochastic Liouville equation (SLE) methodology. Slow reorientations of the whole protein are superimposed on fast chain motions, which have been identified with conformational jumps and fluctuations in the minima of the chain torsional potential. Fast chain motions are introduced in the SLE for the protein reorientations through partially averaged magnetic tensors and relaxation times calculated according to the motional narrowing theory. The 72R1 and 72R2 mutants of T4 lysozyme, which bear the spin label at a solvent-exposed helix site, have been taken as test systems. For the side chain of the R2 spin label, only a few noninterconverting conformers are possible, whose mobility is limited to torsional fluctuations, yielding almost identical spectra, typical of slightly mobile nitroxides. In the case of R1, more complex spectra result from the simultaneous presence of constrained and mobile chain conformers, with relative weights that can depend on the local environment. The model provides an explanation for the experimentally observed dependence of the spectral line shapes on temperature, solvent, and pattern of substituents in the pyrroline ring. The relatively simple methodology presented here allows the introduction of realistic features of the spin probe dynamics into the simulation of ESR spectra of spin-labeled proteins; moreover, it provides suggestions for a proper account of such dynamics in more sophisticated approaches.
|
|
|
Dynamics of the Nitroxide Side Chain in Spin-Labeled Proteins
F. Tombolato, A. Ferrarini, and J.H. Freed.
J. Phys. Chem. B 110, 26248-26259 (2006).
<doi: 10.1021/jp0629487>
PMID:
17181283
PMCID:
PMC2883179
Publication #323
|

|
|
ABSTRACT: The dynamics of the tether linking methanethiosulfonate (MTSSL) spin probes to α-helices has been investigated with the purpose of rationalizing its effects on ESR line shapes. Torsional profiles for the chain bonds have been calculated ab initio, and steric interactions with the α-helix and the neighboring residues have been introduced at the excluded-volume level. As a consequence of the restrictions deriving from chain geometry and local constraints, a limited number of allowed conformers has been identified that undergo torsional oscillations and conformational jumps. Torsional fluctuations are described as damped oscillations, while transition rates between conformers are calculated according to the Langer multidimensional extension of the Kramers theory. The time scale and amplitude of the different motions are compared; the major role played by rotations of the outermost bonds of the side chain emerges, along with the effects of substituents in the pyrroline ring on the conformer distribution and dynamics. The extent and symmetry of magnetic tensor averaging produced by the side chain motions are estimated, the implications for the ESR spectra of spin-labeled proteins are discussed, and suggestions for the introduction of realistic features of the spin probe dynamics into the line shape simulation are presented.
|
|
|
Oxygen Effects on the EPR Signals from Wood Charcoals: Experimental Results and the Development of a Model
O.Y. Grinberg, B.B. Williams, A.E. Ruuge, S.A. Grinberg, D.E. Wilcox, H.M. Swartz, and J.H. Freed.
J. Phys. Chem. B 111, 13316-13324 (2007).
<doi: 10.1021/jp072656l>
PMID:
17973414
Publication #322
|
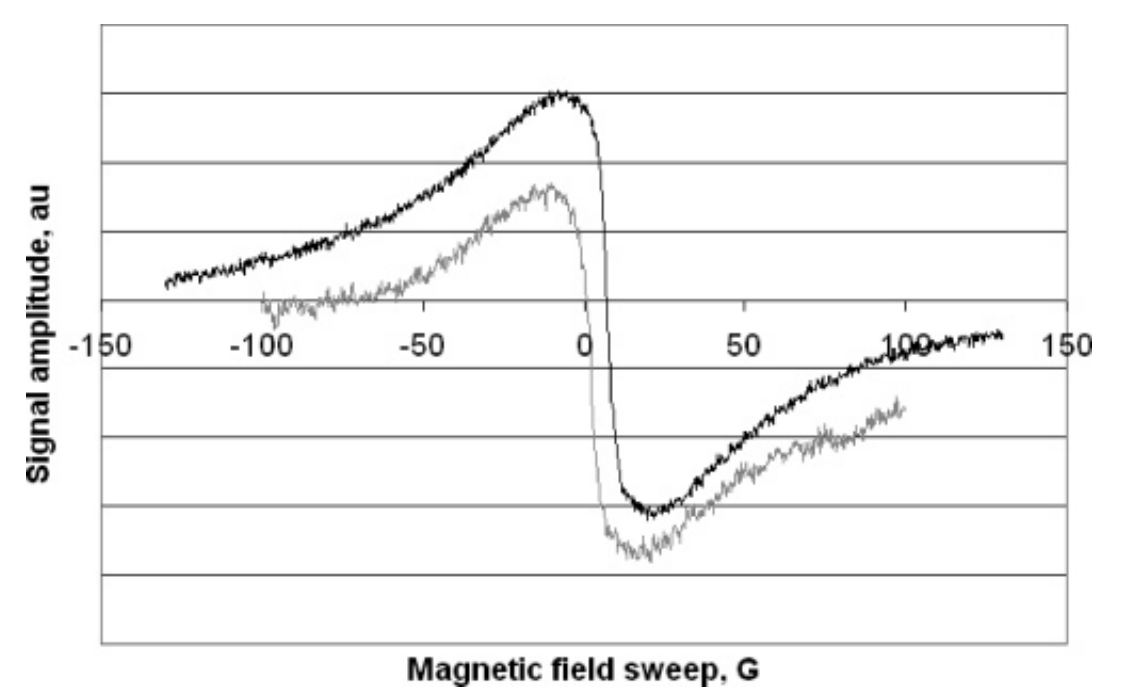
|
|
ABSTRACT: Charcoals prepared from certain tropical woods contain stable paramagnetic centers, and these have been characterized by EPR spectroscopy in the absence and presence of oxygen. The EPR-detectable spin density has been determined, as has been the temperature- and frequency-dependence of the oxygen broadening of the EPR signal, which is orders of magnitude larger than that observed with other materials, such as lithium phthalocyanine. Three Lorentzian components are required to fit the char EPR spectrum in the presence of oxygen, and the oxygen-dependence of the line width, intensity, and resonance position of the three components have been quantified. These results and the properties of porous carbonaceous materials are used to develop a model to explain the effect of oxygen on the char EPR spectral properties. The model is based on oxygen adsorption on the char surface according to a Langmuir isotherm and a dipolar interaction between the paramagnetic adsorbed gas and the charcoal spins. The three EPR components are correlated with the three known classes (sizes) of pores in charcoal, with the largest line broadening attributed to dipolar relaxation of spins in micropores, which have a larger specific surface area and a higher concentration of adsorbed oxygen. An attenuated, but similar, EPR response to oxygen by chars when they are immersed in aqueous solution is attributed to water competition with oxygen for adsorption on the char surface.
|
|
|
Methyl Dynamics in Proteins from NMR Slowly Relaxing Local Structure Spin Relaxation Analysis: A New Perspective
Eva Meirovitch, Antonino Polimeno, and Jack H. Freed.
J. Phys. Chem. B 110, 20615-20628 (2006).
<doi: 10.1021/jp061403+>
PMID:
17034251
Publication #321
|
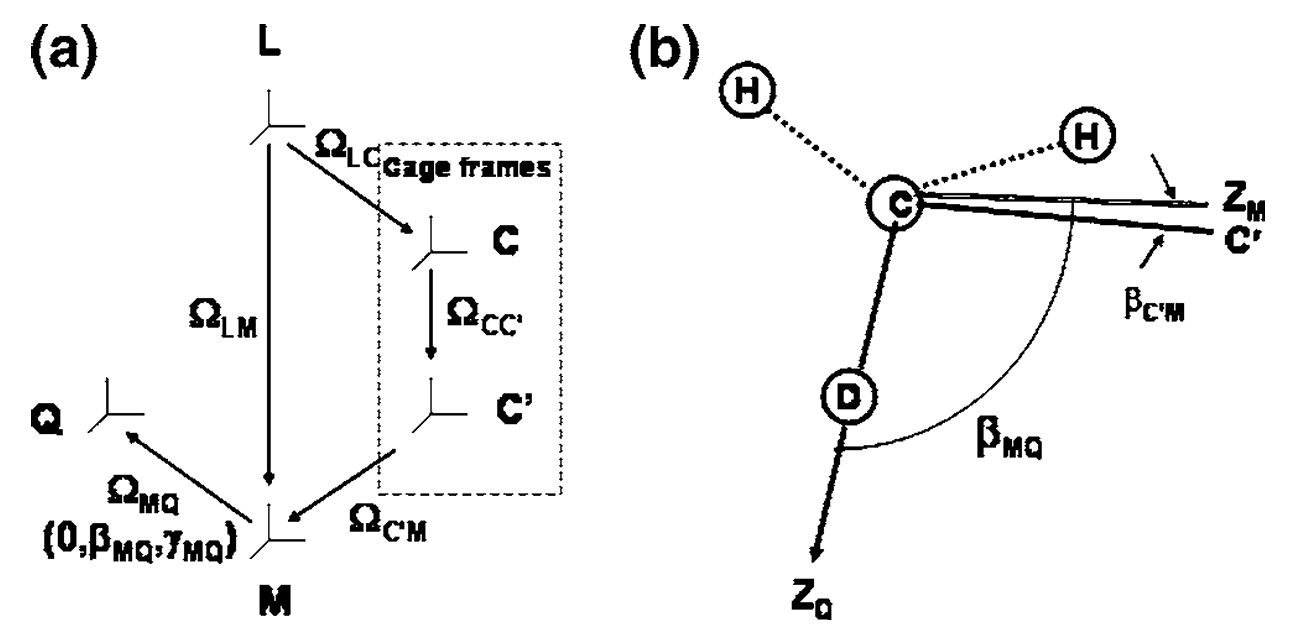
|
|
ABSTRACT: NMR spin relaxation of 2H nuclei in 13CH2D groups is a powerful method for studying side-chain motion in proteins. The analysis is typically carried out with the original model-free (MF) approach adapted to methyl dynamics. The latter is described in terms of axial local motions around, and of, the methyl averaging axis, mutually decoupled and independent of the global motion of the protein. Methyl motion is characterized primarily by the axial squared order parameter, S2axis, associated with fluctuations of the methyl averaging axis. This view is shown to be oversimplified by applying to typical experimental data the slowly relaxing local structure (SRLS) approach of Polimeno and Freed (Adv. Chem. Phys. 1993, 83, 89) which can be considered the generalization of the MF approach. Neglecting mode coupling and the asymmetry of the local ordering and treating approximately features of local geometry imply inaccurate values of S2axis, hence of the residual configurational entropy derived from it. S2axis, interpreted as amplitude of motion, was found to range from near disorder to almost complete order. Contrary to this picture, we find with the SRLS approach a moderate distribution in the magnitude of asymmetric local ordering and significant variation in its symmetry. The latter important property can be associated implicitly with the contribution of side-chain rotamer jumps. This is consistent with experimental residual dipolar coupling studies and theoretical work based on molecular dynamics simulations and molecular mechanics considerations. Configurational entropy is obtained in the SRLS approach directly from experimentally determined asymmetric potentials. Inconsistency between order parameters from 2H relaxation and from ηHC-HH cross-correlation and increase in order parameters with increasing temperature were observed with the MF approach. These discrepancies are reconciled, and physically tenable temperature dependence is obtained with the SRLS approach.
|
|
|
Inter-Helix Distances in Lysophospholipid Micelle-Bound α-Synuclein from Pulsed ESR Measurements
P. Borbat, T.F. Ramlall, J.H. Freed, and D. Eliezer.
J. Am. Chem. Soc. 128, 10004-05 (2006).
Supporting Information
<doi: 10.1021/ja063122l>
PMID:
16881616
Publication #320
|
|
|
ABSTRACT: We demonstrate the use of pulsed ESR spectroscopy to measure intramolecular distances in the Parkinson's disease-associated protein α-synuclein bound to detergent and lysophospholipid micelles. We show that the inter-helical separation between the two helices formed upon binding to micelles is dependent on micelle composition, with micelles formed from longer acyl chains leading to an increased splaying of the two helices. Our data suggest that the topology of α-synuclein is not strongly constrained by the linker region between the two helices and instead depends on the geometry of the surface to which the protein is bound.
|
|
|
Protein Dynamics from NMR: The Slowly Relaxing Local Structure Analysis Compared with Model-Free Analysis
E. Meirovitch, Y.E. Shapiro, A. Polimeno, and J.H. Freed.
J. Phys. Chem. A 110, 8366-8396 (2006).
<doi: 10.1021/jp056975t>
PMID:
16821820
PMCID:
PMC2758167
Publication #319
|
|
|
ABSTRACT: 15N−1H spin relaxation is a powerful method for deriving information on protein dynamics. The traditional method of data analysis is model-free (MF), where the global and local N−H motions are independent and the local geometry is simplified. The common MF analysis consists of fitting single-field data. The results are typically field-dependent, and multifield data cannot be fit with standard fitting schemes. Cases where known functional dynamics has not been detected by MF were identified by us and others. Recently we applied to spin relaxation in proteins the slowly relaxing local structure (SRLS) approach, which accounts rigorously for mode mixing and general features of local geometry. SRLS was shown to yield MF in appropriate asymptotic limits. We found that the experimental spectral density corresponds quite well to the SRLS spectral density. The MF formulas are often used outside of their validity ranges, allowing small data sets to be force-fitted with good statistics but inaccurate best-fit parameters. This paper focuses on the mechanism of force-fitting and its implications. It is shown that MF analysis force-fits the experimental data because mode mixing, the rhombic symmetry of the local ordering and general features of local geometry are not accounted for. Combined multifield multitemperature data analyzed with the MF approach may lead to the detection of incorrect phenomena, and conformational entropy derived from MF order parameters may be highly inaccurate. On the other hand, fitting to more appropriate models can yield consistent physically insightful information. This requires that the complexity of the theoretical spectral densities matches the integrity of the experimental data. As shown herein, the SRLS spectral densities comply with this requirement.
|
|
|
ESR Microscopy and Nanoscopy with "Induction" Detection
A. Blank and J.H. Freed.
Isr. J. Chem. 46, 423-438 (2006).
<doi: 10.1560/IJC_46_4_423>
Publication #318
|

|
|
ABSTRACT: The current state-of-the-art in the fields of Nuclear Magnetic Resonance (NMR) and Electron Spin Resonance (ESR) micro-imaging is reviewed. Special attention is given to the uniqueness and the advantages of the conventional "induction" detection method with respect to other emerging sensitive magnetic resonance detection and imaging techniques. Following this, a theoretical description of the factors affecting the sensitivity and resolution in induction detection ESR is provided. Based on the theory, new approaches to substantially improve ESR capabilities, both at room temperature and at low cryogenic temperatures, are discussed. Representative results of experiments conducted at room temperature and at frequencies of 10–16 GHz show that with test samples, a sensitivity of ˜107 electron spins and a resolution of ˜3 microns are currently available. The results confirm the validity of the theoretical approach and confirm the value of striving for even higher frequencies and lower temperatures, in order to further improve the performance Finally, some of the current and potential applications of ESR microscopy and nanoscopy (involving imaging with a resolution of ˜100 nm or better) are presented.
|
|
|
Two-Pulse Electron Spin Echo Study of Deuterium-Helium Solids
E.P. Bernard, V.V. Khmelenko, E. Vehmanen, P.P. Borbat, J.H. Freed, and D.M. Lee.
Proc. 24th Intl. Conf. on Low Temp. Physics, 372-373 (2006).
<doi: 10.1063/1.2354742>
Publication #317
|
|
|
ABSTRACT: We measured electron spin echoes from deuterium atoms within deuterium-helium solids with an X-band pulse ESR spectrometer. Our two-pulse electron spin echo envelope modulation (ESEEM) measurements are well described by a model that places 50% – 60% of the deuterium atoms at the interface between the molecular deuterium nanoclusters and the superfluid liquid helium. The remainder of the atoms lie in substitutional sites within the nanoclusters. We also report the spin-lattice relaxation times T1 for these atoms.
|
|
|
Pulse Electron Spin Resonance Method for Investigation of Atoms in Impurity-Helium Solids
E.P. Bernard, V.V. Khmelenko, E. Vehmanen, P.P. Borbat, J.H. Freed, and D.M. Lee.
Proc. 24th Intl. Conf. on Low Temp. Physics, 1659-1660 (2006).
<doi: 10.1063/1.2355345>
Publication #316
|
|
|
ABSTRACT: We have constructed an X-band pulse electron spin resonance spectrometer for the investigation of hydrogen, deuterium, and nitrogen impurity-helium solids at low temperatures. The spectrometer is of a conventional homodyne detection design incorporating a sampling oscilloscope but uses a modified CW ESR resonant cavity.
|
|
|
High-Field ESR on Aligned Membranes: A Simple Method to Record Spectra from Different Membrane Orientations in the Magnetic Field
B. Dzikovski, K. Earle, S. Pachtchenko, J. Freed.
J. Magn. Reson. 179, 273-279 (2006).
<doi: 10.1016/j.jmr.2005.12.015>
PMID:
16427793
Publication #315
|
|
|
ABSTRACT: A combination of isopotential spin-dry ultracentrifugation (ISDU) and microtome techniques was used to facilitate the collection of high field/high frequency (170 GHz) ESR spectra corresponding to different orientations of the membrane normal relative to the magnetic field. This technique is particularly valuable for aligned biological samples in vitro. At 170 GHz, conventional sample preparation techniques based solely on ISDU constrained the sample to be oriented so that the membrane normal was parallel to the applied magnetic field due to the geometry and the millimeter wave field distribution of the Fabry–Pérot resonator used in these experiments. This orientational constraint limited the information that could be obtained from aligned membranes at high field. The combined ISDU/microtome technique overcame this limitation. Spectra from spin-labeled Gramicidin A and the spin label cholestane in aligned DPPC membranes provide a demonstration of the technique. We also discuss some virtues of high field/high frequency ESR on aligned membranes compared to X-band.
|
|
|
Reconstruction of the Chemotaxis Receptor-Kinase Assembly
S.-Y. Park, P.P. Borbat, G. Gonzalez-Bonet, J. Bhatnagar, A.M. Pollard, J.H. Freed, A.M. Bilwes & B.R. Crane.
Nat. Struct. Molec. Biol. 13, 400-407 (2006).
Supporting Information
<doi: 10.1038/nsmb1085>
PMID:
16622408
Publication #314
|
|
|
ABSTRACT: In bacterial chemotaxis, an assembly of transmembrane receptors, the CheA histidine kinase and the adaptor protein CheW processes environmental stimuli to regulate motility. The structure of a Thermotoga maritima receptor cytoplasmic domain defines CheA interaction regions and metal ion–coordinating charge centers that undergo chemical modification to tune receptor response. Dimeric CheA–CheW, defined by crystallography and pulsed ESR, positions two CheWs to form a cleft that is lined with residues important for receptor interactions and sized to clamp one receptor dimer. CheW residues involved in kinase activation map to interfaces that orient the CheW clamps. CheA regulatory domains associate in crystals through conserved hydrophobic surfaces. Such CheA self-contacts align the CheW receptor clamps for binding receptor tips. Linking layers of ternary complexes with close-packed receptors generates a lattice with reasonable component ratios, cooperative interactions among receptors and accessible sites for modification enzymes.
|
|
|
Electron Spin Resonance Microscopy Applied to the Study of Controlled Drug Release
A. Blank, J.H. Freed, N.P. Kumar, C.-H. Wang.
J. Cont. Rel. 111, 174-184 (2006).
<doi: 10.1016/j.jconrel.2005.11.019>
PMID:
16460828
Publication #313
|
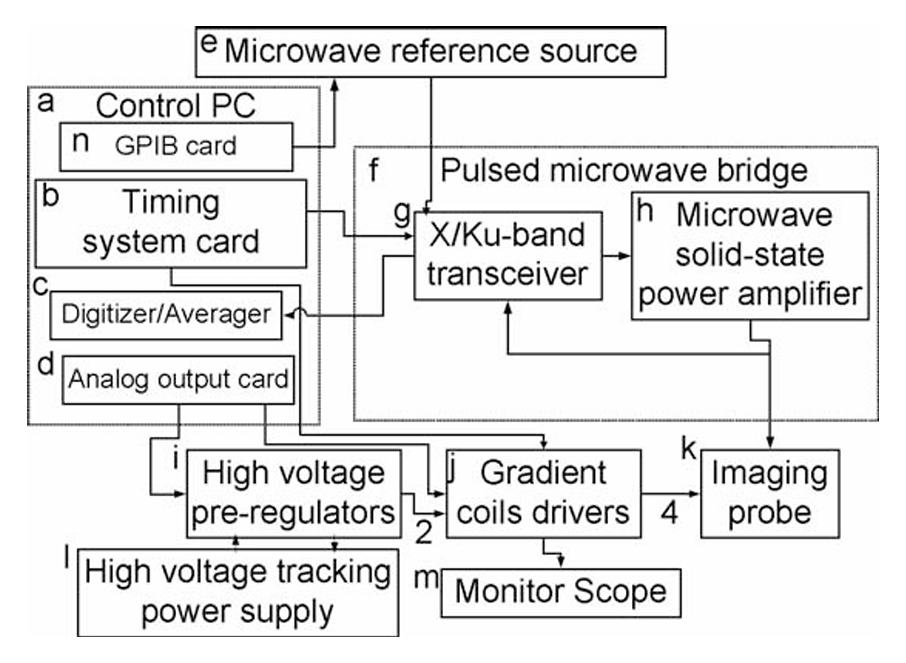
|
|
ABSTRACT: We describe our recent developments towards 3D micron-scale imaging capability, based on electron spin resonance (ESR), and its application to the study of controlled release. The method, termed ESR microscopy (ESRM), is an extension of the conventional "millimeter-scale" ESR imaging technique. It employs paramagnetic molecules (such as stable radicals or spin-labeled drugs) and may enable one to obtain accurate 3D spatially resolved information about the drug concentration, its self-diffusion tensor, rotational correlation time and the pH in the release matrix. Theoretical calculations, along with initial experimental results, suggest that a 3D resolution of ˜ 1 μm is feasible with this method. Here we were able to image successfully a high spin concentration sample with a resolution of ˜ 3 × 3 × 8 μm and subsequently study a single ˜ 120 μm biodegradable microsphere, internalized with a dilute solution of trityl radical, with a resolution of ˜ 12.7 × 13.2 × 26 μm. Analysis of the microsphere ESR imaging data revealed a likely increase in the viscosity inside the sphere and/or binding of the radical molecule to the sphere matrix. Future directions for progress are also discussed.
|
|
|
EPR Distance Measurements Support a Model for Long-Range Radical Initiation in E. coli Ribonucleotide Reductase
M. Bennati, J.H. Robblee, V. Mugnaini, J. Stubbe, J.H. Freed, and P. Borbat.
J. Am. Chem. Soc. 127, 15014-15015 (2005).
<doi: 10.1021/ja054991y>
PMID:
16248626
Publication #312
|
|
|
ABSTRACT: The class I E. coli ribonucleotide reductase, composed of homodimers of R1 and R2, catalyzes the conversion of nucleoside diphosphates to deoxynucleoside diphosphates. The reduction process involves the tyrosyl radical on R2 that generates a transient thiyl radical on R1 over a proposed distance of 35 Å. A mechanism-based inhibitor, 2′-azido-2′-deoxyuridine-5′-diphosphate, that reduces the tyrosyl radical on R2 and forms a nitrogen-centered radical on R1 has provided a method to measure the diagonal distance between the two subunits. PELDOR and DQC paramagnetic resonance methods give rise to a distance of 48 Å, similar to that calculated from a docking model of the R1 and R2 structures.
|
|
|
Coexisting Domains in the Plasma Membranes of Live Cells Characterized by ESR Spectroscopy
M.J. Swamy, L. Ciani, M. Ge, A.K. Smith, D. Holowka, B. Baird, and J.H. Freed.
Biophys. J. 90, 4452-4465 (2006).
Supporting Information
<doi: 10.1529/biophysj.105.070839>
PMID:
16565045
PMCID:
PMC1471862
Publication #311
|
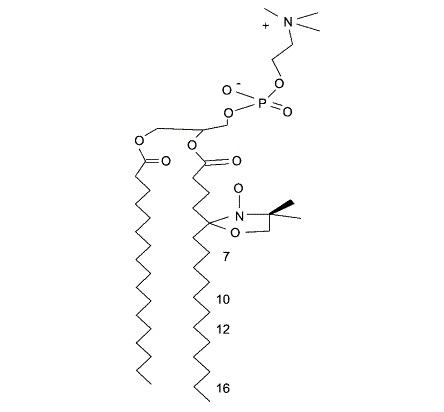
|
|
ABSTRACT: The importance of membrane-based compartmentalization in eukaryotic cell function has become broadly appreciated, and a number of studies indicate that these eukaryotic cell membranes contain coexisting liquid-ordered (Lo) and liquid-disordered (Ld) lipid domains. However, the current evidence for such phase separation is indirect, and so far there has been no direct demonstration of differences in the ordering and dynamics for the lipids in these two types of regions or their relative amounts in the plasma membranes of live cells. In this study, we provide direct evidence for the presence of two different types of lipid populations in the plasma membranes of live cells from four different cell lines by electron spin resonance. Analysis of the electron spin resonance spectra recorded over a range of temperatures, from 5 to 37°C, shows that the spin-labeled phospholipids incorporated experience two types of environments, Lo and Ld, with distinct order parameters and rotational diffusion coefficients but with some differences among the four cell lines. These results suggest that coexistence of lipid domains that differ significantly in their dynamic order in the plasma membrane is a general phenomenon. The Lo region is found to be a major component in contrast to a model in which small liquid-ordered lipid rafts exist in a 'sea' of disordered lipids. The results on ordering and dynamics for the live cells are also compared with those from model membranes exhibiting coexisting Lo and Ld phases.
|
|
|
Effects of Finite Pulse Width on Two-Dimensional Fourier Transform Electron Spin Resonance
Z. Liang, R.H. Crepeau, J.H. Freed.
J. Magn. Reson. 177, 247-260 (2005).
<doi: 10.1016/j.jmr.2005.07.024>
PMID:
16150620
Publication #310
|
|
|
ABSTRACT: Two-dimensional (2D) Fourier transform ESR techniques, such as 2D-ELDOR, have considerably improved the resolution of ESR in studies of molecular dynamics in complex fluids such as liquid crystals and membrane vesicles and in spin labeled polymers and peptides. A well-developed theory based on the stochastic Liouville equation (SLE) has been successfully employed to analyze these experiments. However, one fundamental assumption has been utilized to simplify the complex analysis, viz. the pulses have been treated as ideal non-selective ones, which therefore provide uniform irradiation of the whole spectrum. In actual experiments, the pulses are of finite width causing deviations from the theoretical predictions, a problem that is exacerbated by experiments performed at higher frequencies. In the present paper we provide a method to deal with the full SLE including the explicit role of the molecular dynamics, the spin Hamiltonian and the radiation field during the pulse. The computations are rendered more manageable by utilizing the Trotter formula, which is adapted to handle this SLE in what we call a "Split Super-Operator" method. Examples are given for different motional regimes, which show how 2D-ELDOR spectra are affected by the finite pulse widths. The theory shows good agreement with 2D-ELDOR experiments performed as a function of pulse width.
|
|
|
Maximum Entropy: A Complement to Tikhonov Regularization for Determination of Pair Distance Distributions by Pulsed ESR
Y.-W. Chiang, P.P. Borbat, J.H. Freed.
J. Magn. Reson. 177, 184-196 (2005).
<doi: 10.1016/j.jmr.2005.07.021>
PMID:
16137901
Publication #309
|
|
|
ABSTRACT: Tikhonov regularization (TIKR) has been demonstrated as a powerful and valuable method for the determination of distance distributions of spin-pairs in bi-labeled biomolecules directly from pulsed ESR signals. TIKR is a direct method, which requires no iteration, and, therefore, provides a rapid and unique solution. However, the distribution obtained tends to exhibit oscillatory excursions with negative portions in the presence of finite noise, especially in the peripheral regions of the distribution. The Shannon–Jaynes entropy of a probability distribution provides an intrinsic non-negativity constraint on the probability distribution and an unbiased way of obtaining information from incomplete data. We describe how the maximum entropy regularization method (MEM) may be applied to solve the ill-posed nature of the dipolar signal in pulsed ESR. We make use of it to suppress the negative excursions of the distance distribution and to increase the tolerance to noise in the dipolar signal. Model studies and experimental data are investigated, and they show that, with the initial or "seed" probability distribution that is required for MEM taken as the TIKR result, then MEM is able to provide a regularized solution, subject to the non-negativity constraint, and it is effective in dealing with noise that is problematic for TIKR. In addition we have incorporated into our MEM method the ability to extract the intermolecular dipolar component, which is embedded in the raw experimental data. We find that MEM minimization, which is implemented iteratively, is greatly accelerated using the TIKR result as the seed, and it converges more successfully. Thus we regard the MEM method as a complement to TIKR by securing a positive pair distance distribution and enhancing the accuracy of TIKR.
|
|
|
High-Frequency ESR at ACERT
K.A. Earle, B. Dzikovski, W. Hofbauer, J.K. Moscicki and J.H. Freed.
Magn. Reson. Chem. 43, S256-S266 (2005).
|
|
|
High-Frequency ESR at ACERT
K.A. Earle, B. Dzikovski, W. Hofbauer, J.K. Moscicki and J.H. Freed.
Magn. Reson. Chem. 43, S256-S266 (2005).
<doi: 10.1002/mrc.1684>
PMID:
16235203
Publication #308
|

|
|
ABSTRACT: High-field ESR offers many advantages in exploring fundamental questions of structure and dynamics in chemical, biological and physical samples. We provide a review of recent work performed at ACERT demonstrating the utility and flexibility of our methods for extracting both qualitative and quantitative information from a variety of systems. In particular, we emphasize the utility of multi-frequency ESR techniques for unraveling the details of the complex dynamical modes of proteins in solution and in heterogeneous systems such as lipid bilayers. We also include indications of directions for future work where appropriate.
|
|
|
A Variable Temperature EPR Study of Mn2+-doped NH4Cl0.9I0.1 Single Crystal at 170 GHz: Zero-Field Splitting Parameter and its Absolute Sign
S.K. Misra, S.I. Andronenko, P. Chand, K.A. Earle, S.V. Paschenko, J.H. Freed.
J. Magn. Reson. 174, 265-269 (2005).
<doi: 10.1016/j.jmr.2005.02.015>
PMID:
15862243
Publication #307
|
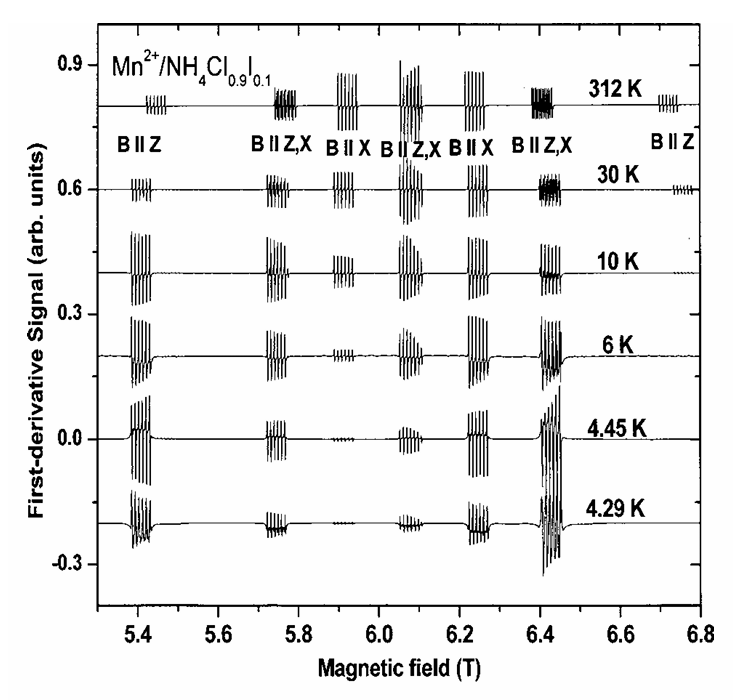
|
|
ABSTRACT: EPR measurements have been carried out on a single crystal of Mn2+-doped NH4Cl0.9I0.1 at 170-GHz in the temperature range of 312–4.2 K. The spectra have been analyzed (i) to estimate the spin-Hamiltonian parameters; (ii) to study the temperature variation of the zero-field splitting (ZFS) parameter; (iii) to confirm the negative absolute sign of the ZFS parameter unequivocally from the temperature-dependent relative intensities of hyperfine sextets at temperatures below 10 K; and (iv) to detect the occurrence of a structural phase transition at 4.35 K from the change in the structure of the EPR lines with temperature below 10 K.
|
|
|
Pulsed Three-Dimensional Electron Spin Resonance Microscopy
A. Blank, C.R. Dunnam, P.P. Borbat, and J.H. Freed.
Appl. Phys. Lett. 85, 5430-5432 (2004).
<doi: 10.1063/1.1828599>
Publication #306
|
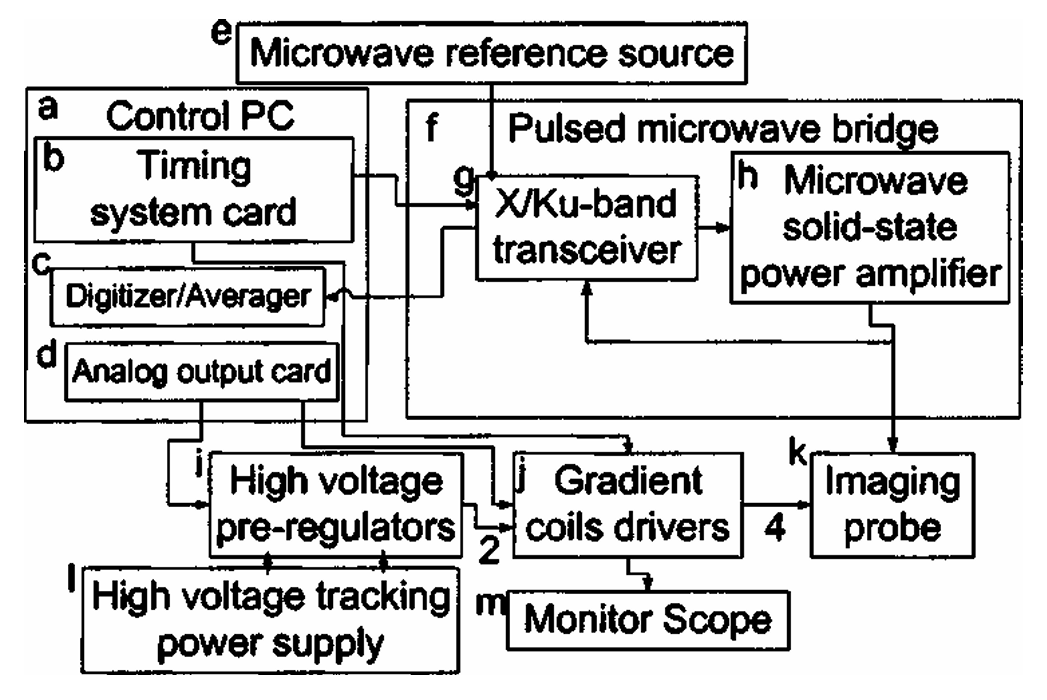
|
|
ABSTRACT: A three-dimensional (3D) electron spin resonance (ESR) microimaging system, operating in pulse mode at 9 GHz is presented. This microscope enables the acquisition of spatially resolved magnetic resonance signals of free-radicals in solid or liquid samples with a resolution of up to ˜3.5 × 7 × 11.4 μm in 20 min of acquisition. The detection sensitivity at room temperature is ˜1.2 × 109 spins / √Hz, which enables the measurement of ˜2 × 107 spins in each voxel after 60 min of acquisition. The resolution and detection sensitivity are the best obtained so far for ESR at ambient conditions of temperature and pressure. This ESR microscope can be employed in the investigation of a variety of samples in the fields of botany, life sciences, and materials science.
|
|
|
The Determination of Pair Distance Distributions by Pulsed ESR Using Tikhonov Regularization
Y.-W. Chiang, P.P. Borbat, J.H. Freed.
J. Magn. Reson. 172, 279-295 (2005).
<doi: 10.1016/j.jmr.2004.10.012>
PMID:
15649755
Publication #305
|
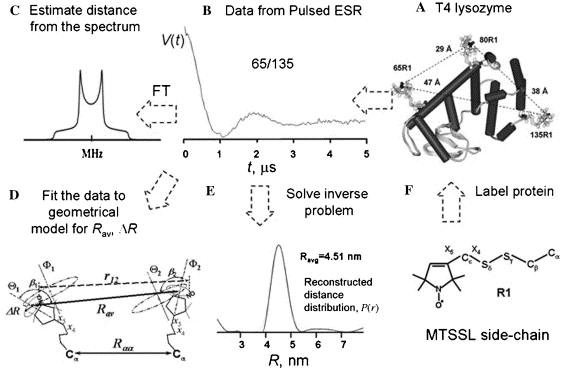
|
|
ABSTRACT: Pulsed ESR techniques with the aid of site-directed spin labeling have proven useful in providing unique structural information about proteins. The determination of distance distributions in electron spin pairs directly from the dipolar time evolution of the pulsed ESR signals by means of the Tikhonov regularization method is reported. The difficulties connected with numerically inverting this ill-posed mathematical problem are clearly illustrated. The Tikhonov regularization with the regularization parameter determined by the L-curve criterion is then described and tested to confirm its accuracy and reliability. The method is applied to recent experimental results on doubly labeled proteins that have been studied using two pulsed ESR techniques, double quantum coherence (DQC) ESR and double electron–electron resonance (DEER). The extracted distance distributions are able to provide valuable information about the conformational constraints in various partially folded states of proteins. This study supplies a mathematically reliable method for extracting pair distributions from pulsed ESR experimental data and has extended the use of pulsed ESR to provide results of greater value for structural biology.
|
|
|
New Method for Determining Tie-lines in Coexisting Membrane Phases using Spin-label ESR
Y.-W. Chiang, J. Zhao, J. Wu, Y. Shimoyama, J.H. Freed, and G.W. Feigenson.
Biochim. Biophys. Acta 1668, 99-105 (2005).
<doi: 10.1016/j.bbamem.2004.11.010>
PMID:
15670735
Publication #304
|
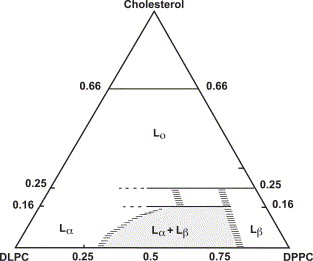
|
|
ABSTRACT: A full description of coexisting phases includes their respective compositions, which are provided by the thermodynamic tie-lines. Fluorescence microscopy enables visualization of coexisting lipid phases in giant unilamellar phases, but the composition information is missing. For cholesterol-containing lipid mixtures, knowledge of the compositions of the coexisting phases is important for understanding the nature of "membrane rafts". We propose and demonstrate a new method, based on ESR spectroscopy, for determining tie-lines in regions of two-phase coexistence in a ternary lipid mixture. Over 100 different lipid compositions containing the spin-labeled phospholipid 16-PC in or near the two-phase coexistence region of the liquid-disordered and the gel phases of dipalmitoyl-PC/dilauroyl-PC/cholesterol (DPPC/DLPC/Chol) were studied to determine five tie-lines, spread over virtually the full range of this coexistence region. The method is based on the facts that (1) along a tie-line the ESR spectrum must be a superposition of the two ESR spectra from the respective single phases at the phase boundaries (connected by the tie-line) in a ratio given by the lever rule; (2) along a tie-line the partition coefficient, Kp, for the spin-label, which is also determined in our method, must be constant. We do find that Kp for 16-PC is close to unity, but its value depends on the particular tie-line. The coexisting phases in equilibrium are characterized by the Kp of the spin-label and its respective dynamic parameters obtained from fitting the ESR spectra to dynamical models.
|
|
|
Dynamic Molecular Structure of DPPC-DLPC-Cholesterol Ternary Lipid System by Spin-Label Electron Spin Resonance
Y.-W. Chiang, Y. Shimoyama, G.W. Feigenson, and J.H. Freed.
Biophys. J. 87, 2483-2496 (2004).
<doi: 10.1529/biophysj.104.044438>
PMID:
15454445
PMCID:
PMC1304668
Publication #303
|
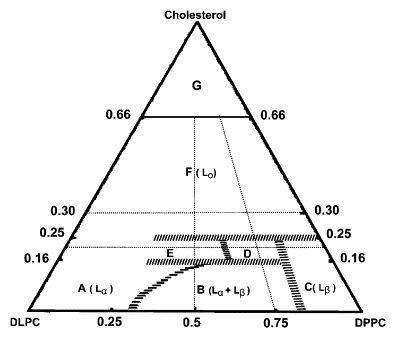
|
|
ABSTRACT: The hydrated ternary lamellar lipid mixture of dipalmitoyl-PC/dilauroyl-PC/cholesterol (DPPC/DLPC/Chol) has been studied by electron spin resonance (ESR) to reveal the dynamic structure on a molecular level of the different phases that exist and coexist over virtually the full range of composition. The spectra for more than 100 different compositions at room temperature were analyzed by nonlinear least-squares fitting to provide the rotational diffusion rates and order parameters of the end-chain labeled phospholipid 16-PC. The ESR spectra exhibit substantial variation as a function of composition, even though the respective phases generally differ rather modestly from each other. The Lα and Lβ phases are clearly distinguished, with the former exhibiting substantially lower ordering and greater motional rates, whereas the well-defined LOphase exhibits the greatest ordering and relatively fast motional rates. Typically, smaller variations occur within a given phase. The ESR spectral analysis also yields phase boundaries and coexistence regions which are found to be consistent with previous results from fluorescence methods, although new features are found. Phase coexistence regions were in some cases confirmed by observing the existence of isosbestic points in the absorption mode ESR spectra from the phases. The dynamic structural properties of the DPPC-rich Lβ and DLPC-rich Lα phases, within their two-phase coexistence region do not change with composition along a tie-line, but the ratio of the two phases follows the lever rule in accordance with thermodynamic principles. The analysis shows that 16-PC spin-label partitions nearly equally between the Lα and Lβ phases, making it a useful probe for studying such coexisting phases. Extensive study of two-phase coexistence regions requires the determination of tie-lines, which were approximated in this study. However, a method is suggested to accurately determine the tie-lines by ESR.
|
|
|
Spin-Labeled Gramicidin A: Channel Formation and Dissociation
B.G. Dzikovski, P.P. Borbat, and J.H. Freed.
Biophys. J. 87, 3504-3517 (2004).
<doi: 10.1529/biophysj.104.044305>
PMID:
15326023
PMCID:
PMC1304816
Publication #302
|
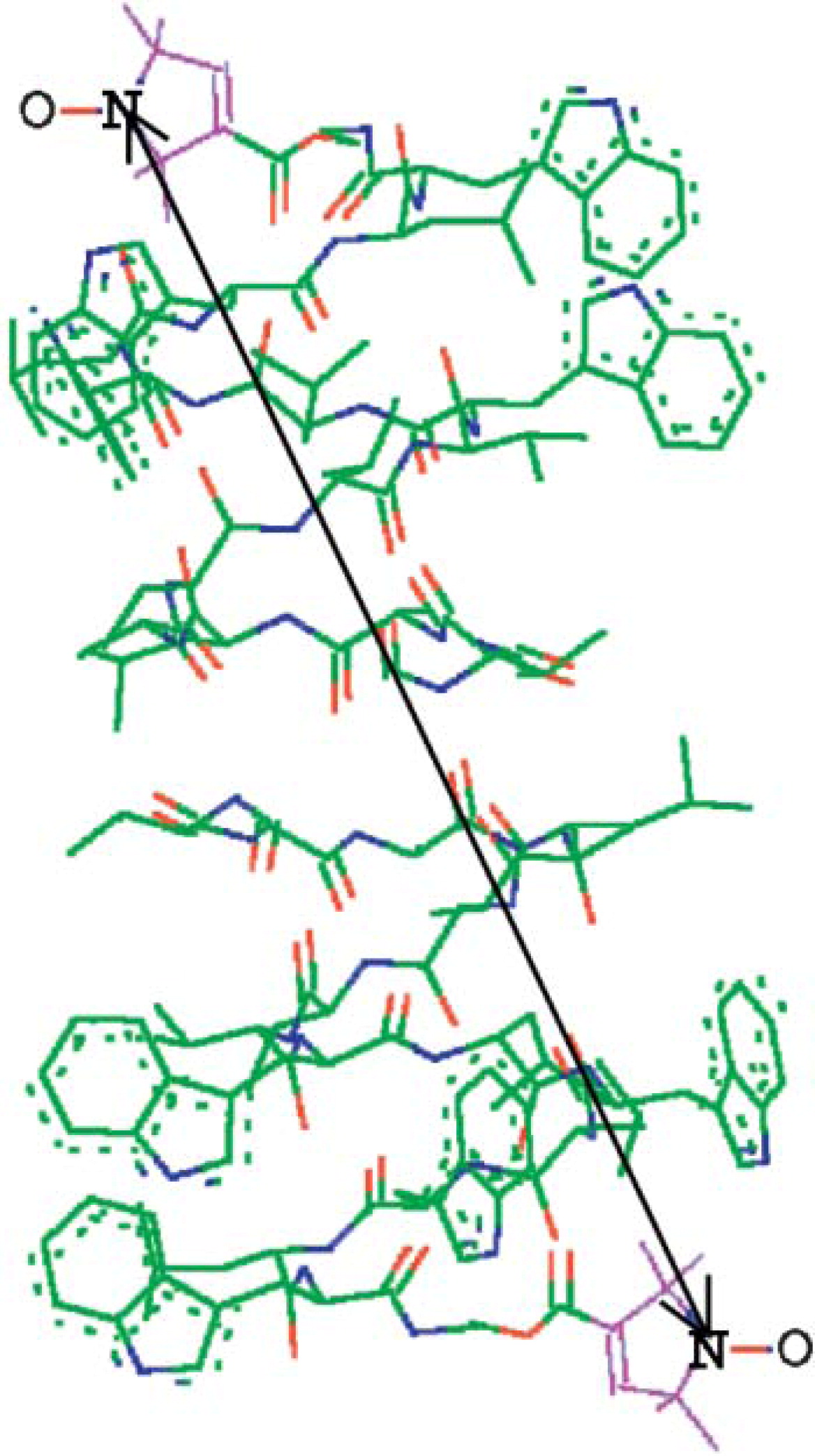
|
|
ABSTRACT: Gramicidin A was studied by continuous wave electron spin resonance (CW-ESR) and by double-quantum coherence electron spin resonance (DQC-ESR) in several lipid membranes (using samples that were macroscopically aligned by isopotential spin-dry ultracentrifugation) and vesicles. As a reporter group, the nitroxide spin-label was attached at the C-terminus yielding the spin-labeled product (GAsl). ESR spectra of aligned membranes containing GAsl show strong orientation dependence. In DPPC and DSPC membranes at room temperature the spectral shape is consistent with high ordering, which, in conjunction with the observed high polarity of the environment of the nitroxide, is interpreted in terms of the nitroxide moiety being close to the membrane surface. In contrast, spectra of GAsl in DMPC membranes indicate deeper embedding and tilt of the NO group. The GAsl spectrum in the DPPC membrane at 35°C (the gel to Pβ phase transition) exhibits sharp changes, and above this temperature becomes similar to that of DMPC. The dipolar spectrum from DQC-ESR clearly indicates the presence of pairs in DMPC membranes. This is not the case for DPPC, rapidly frozen from the gel phase; however, there are hints of aggregation. The interspin distance in the pairs is 30.9 Å, in good agreement with estimates for the head-to-head GAsl dimer (the channel-forming conformation), which matches the hydrophobic thickness of the DMPC bilayer. Both DPPC and DSPC, apparently as a result of hydrophobic mismatch between the dimer length and bilayer thickness, do not favor the channel formation in the gel phase. In the Pβ and Lα phases of DPPC (above 35°C) the channel dimer forms, as evidenced by the DQC-ESR dipolar spectrum after rapid freezing. It is associated with a lateral expansion of lipid molecules and a concomitant decrease in bilayer thickness, which reduces the hydrophobic mismatch. A comparison with studies of dimer formation by other physical techniques indicates the desirability of using low concentrations of GA (˜0.4–1 mol %) accessible to the ESR methods employed in the study, since this yields non-interacting dimer channels.
|
|
|
A Multifrequency Electron Spin Resonance Study of T4 Lysozyme Dynamics Using the Slowly Relaxing Local Structure Model
Z. Liang, Y. Lou, J.H. Freed, L. Columbus and W.L. Hubbell.
J. Phys. Chem. B 108, 17649-17659 (2004).
<doi: 10.1021/jp0484837>
Publication #301
|
|
|
ABSTRACT: Electron spin resonance (ESR) spectra were obtained at 250 and 9 GHz for nitroxide-labeled mutants of the protein T4 lysozyme in aqueous solution over a range of temperatures from 2 to 37.5 °C. Two mutants labeled at sites 72 and 131 were studied and compared. The mutant sites are solvent exposed and free of tertiary interactions with other side chains, but the former is at the center of a 5 turn helix, whereas the latter site is on a small two and a half turn helix. The 250 GHz ESR spectra, because of their "fast time scale", are rather insensitive to the slow overall tumbling motion of the protein. Thus, they are qualitatively different for the two mutants, implying that there are different local dynamics at the two sites. The 9 GHz spectra, which are significantly affected by the overall tumbling and are less sensitive to the internal dynamics, do not show such marked differences between the two sites. The 250 and 9 GHz spectra for each mutant and temperature were simultaneously fit to the slowly relaxing local structure (SRLS) model for slow-motional ESR, using newly developed software. The SRLS model explicitly accounts for the overall tumbling of the protein and the internal modes of motion, which include the motion of the nitroxide side chain (expected to be the same for both mutants) and backbone fluctuations. Very good simultaneous fits are obtained. Whereas two conformers (or spectral components) are typically detected at the lower temperatures, only a single component is observed at the higher temperatures. The significant differences in the high-frequency spectra for the two mutants are readily attributed mainly to a difference in their respective local ordering. That is, site 72 exhibits significantly greater local ordering than does site 131, which is expected from the greater rigidity of the larger helix on which the 72 site is located. The results of this multifrequency study are compared with a previous 9 GHz study. A description of the application of the SRLS model in such a multifrequency study is provided.
|
|
|
Enrichment of Endoplasmic Reticulum with Cholesterol Inhibits Sarcoplasmic-Endoplasmic Reticulum Calcium ATPase-2b Activity in Parallel with Increased Order of Membrane Lipids
Y. Li, M. Ge, L. Ciani, G. Kuriakose, E.J. Westover, M. Dura, D.F. Covey, J.H. Freed, F.R. Maxfield, J. Lytton, and I. Tabas.
J. Biol. Chem. 279, 37030-37039 (2004).
|
|
|
Enrichment of Endoplasmic Reticulum with Cholesterol Inhibits Sarcoplasmic-Endoplasmic Reticulum Calcium ATPase-2b Activity in Parallel with Increased Order of Membrane Lipids
Y. Li, M. Ge, L. Ciani, G. Kuriakose, E.J. Westover, M. Dura, D.F. Covey, J.H. Freed, F.R. Maxfield, J. Lytton, and I. Tabas.
J. Biol. Chem. 279, 37030-37039 (2004).
<doi: 10.1074/jbc.M405195200>
PMID:
15215242
Publication #300
|

|
|
ABSTRACT: Macrophages in advanced atherosclerotic lesions accumulate large amounts of unesterified, or "free," cholesterol (FC). FC accumulation induces macrophage apoptosis, which likely contributes to plaque destabilization. Apoptosis is triggered by the enrichment of the endoplasmic reticulum (ER) with FC, resulting in depletion of ER calcium stores, and induction of the unfolded protein response. To explain the mechanism of ER calcium depletion, we hypothesized that FC enrichment of the normally cholesterol-poor ER membrane inhibits the macrophage ER calcium pump, sarcoplasmic-endoplasmic reticulum calcium ATPase-2b (SERCA2b). FC enrichment of ER membranes to a level similar to that occuring in vivo inhibited both the ATPase activity and calcium sequestration function of SERCA2b. Enrichment of ER with ent-cholesterol or 14:0-18:0 phosphatidylcholine, which possess the membrane-ordering properties of cholesterol, also inhibited SERCA2b. Moreover, at various levels of FC enrichment of ER membranes, there was a very close correlation between increasing membrane lipid order, as monitored by 16-doxyl-phosphatidycholine electron spin resonance, and SERCA2b inhibition. In view of these data, we speculate that SERCA2b, a conformationally active protein with 11 membrane-spanning regions, loses function due to decreased conformational freedom in FC-ordered membranes. This biophysical model may underlie the critical connection between excess cholesterol, unfolded protein response induction, macrophage death, and plaque destabilization in advanced atherosclerosis.
|
|
|
Measurement of Large Distances in Biomolecules Using Double-Quantum Filtered Refocused Electron Spin-Echoes
P.P. Borbat, J.H. Davis, S.E. Butcher, and J.H. Freed.
J. Am. Chem. Soc. 126, 7746-7747 (2004).
<doi: 10.1021/ja049372o>
PMID:
15212500
Publication #299
|
|
|
ABSTRACT: It is shown how the new technique of double-quantum filtered refocused electron spin–echoes is a significant improvement over double-quantum coherence ESR, since it increases the experimental acquisition time. This enables the measurement of longer distances in bilabeled biomolecules. The method is demonstrated on a long double-stranded A-type RNA, spin labeled at both ends. The measured distance of 72 Å is in excellent agreement with molecular modeling.
|
|
|
A Three-Dimensional Electron Spin Resonance Microscope
A. Blank, C.R. Dunnam, P.P. Borbat, and J.H. Freed.
Rev. Sci. Instrum. 75, 3050-3061 (2004).
<doi: 10.1063/1.1786353>
Publication #298
|
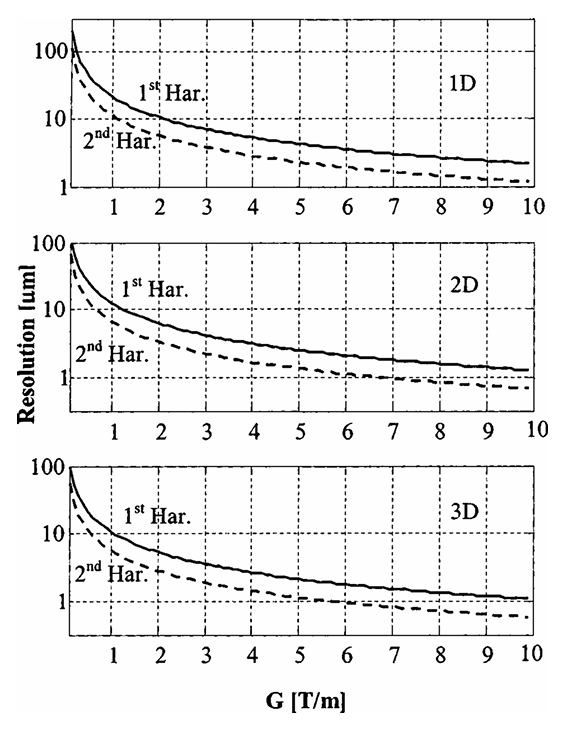
|
|
ABSTRACT: An electron spin resonance (ESR) imaging system, capable of acquiring three-dimensional (3D) images with a resolution of ˜10 × 10 × 30 μm in a few minutes of acquisition, is presented. This ESR microscope employs a commercial continuous wave ESR spectrometer, working at 9.1 GHz, in conjunction with a miniature imaging probe (resonator + gradient coils), gradient current drivers, and control software. The system can acquire the image of a small (˜ 1.5 × 1.5 × 0.25 mm) sample either by the modulated field gradient method, the projection reconstruction method, or by a combination of the two. A short discussion regarding the resolution of the modulated field gradient method in two-dimensional (2D) and 3D imaging is given. Detailed descriptions of the various system components are provided, along with several examples of 2D and 3D images that demonstrate the capabilities of the system.
|
|
|
High-Power 95 GHz Pulsed Electron Spin Resonance Spectrometer
W. Hofbauer, K.A. Earle, C.R. Dunnam, J.K. Moscicki, and J.H. Freed.
Rev. Sci. Instrum. 75, 1194-1208 (2004).
<doi: 10.1063/1.1710700>
Publication #297
|
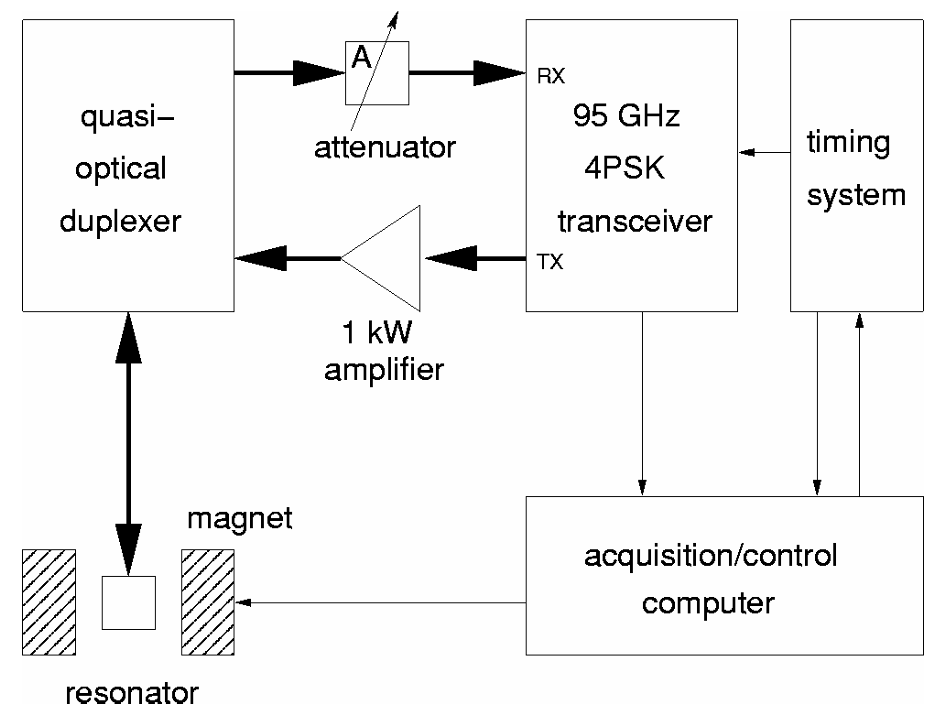
|
|
ABSTRACT: High-field/high-frequency electron spin resonance (ESR) offers improved sensitivity and resolution compared to ESR at conventional fields and frequencies. However, most high-field/high-frequency ESR spectrometers suffer from limited mm-wave power, thereby requiring long mm-wave pulses. This precludes their use when relaxation times are short, e.g., in fluid samples. Low mm-wave power is also a major factor limiting the achievable spectral coverage and thereby the multiplex advantage of Fourier transform ESR (FTESR) experiments. High-power pulses are needed to perform two-dimensional (2D) FTESR experiments, which can unravel the dynamics of a spin system in great detail, making it an excellent tool for studying spin and molecular dynamics. We report on the design and implementation of a high-power, high-bandwidth, pulsed ESR spectrometer operating at 95 GHz. One of the principal design goals was the ability to investigate dynamic processes in aqueous samples at physiological temperatures with the intent to study biological systems. In initial experiments on aqueous samples at room temperature, we achieved 200 MHz spectral coverage at a sensitivity of 1.1×1010√(s) spins and a dead time of less than 50 ns. 2D-electron-electron double resonance experiments on aqueous samples are discussed to demonstrate the practical application of such a spectrometer.
|
|
|
The Development of High Field/High Frequency ESR: Historical Overview
J.H. Freed.
In Very High Frequency (VHF) ESR/EPR, O. Grinberg and L.J. Berliner, Eds.
Biological Magnetic Resonance 22, Chapter 2, pp. 19-43 (2004). (Kluwer, NY).
<doi: 10.1007/978-1-4757-4379-1_2>
Publication #296
|
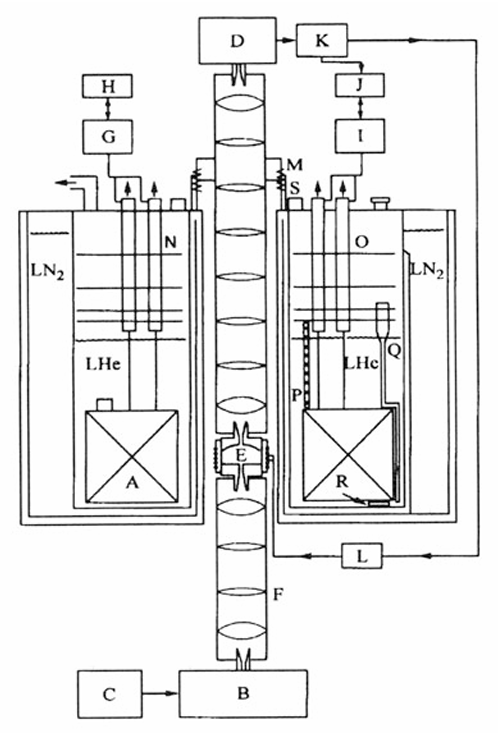
|
|
ABSTRACT: We discuss the development of high field ESR into a powerful and flexible tool for studies of structure and dynamics in a wide variety of systems including those of biological interest. A range of techniques are discussed with particular emphasis on the developments at Cornell University, but the contributions of other groups to the constant refinement of the state of the art are also noted.
|
|
|
High Resolution Electron Spin Resonance Microscopy
A. Blank, C.R. Dunnam, P.P. Borbat, and J.H. Freed.
J. Magn. Reson. 165, 116-127 (2003).
<doi: 10.1016/S1090-7807(03)00254-4>
PMID:
14568522
Publication #295
|
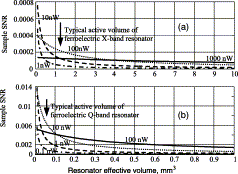
|
|
ABSTRACT: NMR microscopy is routinely employed in fields of science such as biology, botany, and materials science to observe magnetic parameters and transport phenomena in small scale structures. Despite extensive efforts, the resolution of this method is limited (>10 μm for short acquisition times), and thus cannot answer many key questions in these fields. We show, through theoretical prediction and initial experiments, that ESR microscopy, although much less developed, can improve upon the resolution limits of NMR, and successfully undertake the 1 μm resolution challenge. Our theoretical predictions demonstrate that existing ESR technology, along with advanced imaging probe design (resonator and gradient coils), using solutions of narrow linewidth radicals (the trityl family), should yield 64 × 64 pixels 2D images (with z slice selection) with a resolution of 1 × 1 × 10 μm at ˜60 GHz in less than 1 h of acquisition. Our initial imaging results, conducted by CW ESR at X-band, support these theoretical predictions and already improve upon the previously reported state-of-the-art for 2D ESR image resolution achieving ˜10 × 10 μm, in just several minutes of acquisition time. We analyze how future progress, which includes improved resonators, increased frequency of measurement, and advanced pulsed techniques, should achieve the goal of micron resolution.
|
|
|
Mode-Coupling SRLS versus Mode-Decoupled Model-Free N-H Bond Dynamics: Mode-Mixing and Renormalization
E. Meirovitch, Y.E. Shapiro, Z. Liang, and J.H. Freed.
J. Phys. Chem. B 107, 9898-9904 (2003).
<doi: 10.1021/jp030502>
Publication #294
|
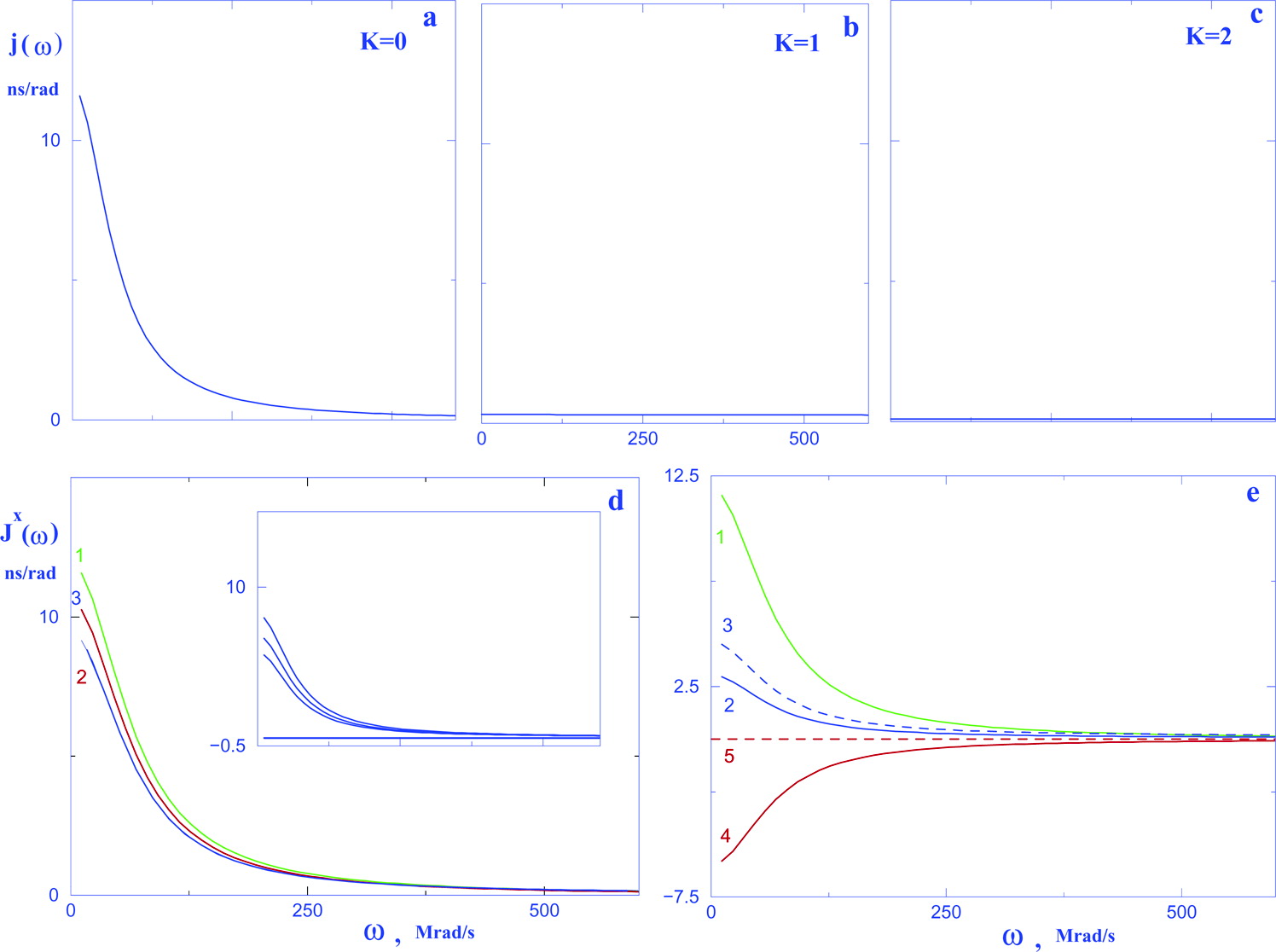
|
|
ABSTRACT: The common approach to N−H motion in proteins is model-free (MF), where the global (RC) and local (RL) motions are assumed decoupled. We have recently applied to N−H bond dynamics the slowly relaxing local structure (SRLS) model, which accounts rigorously for mode-coupling. The original and extended MF formulas are perturbational expansions of SRLS with respect to the local ordering, (S02)2, when RL ≫ RC. Their functional form, number of terms equal to the number of dynamic modes, is implied by mode-decoupling, and the free diffusion eigenvalue, 1∕τ = 6RL, by the absence of strong-potential-induced renormalization. However, for N−H motion, (S02)2 is high and in the extended MF regime RL⊥ ≈ RC. Although the functional form of the original MF formula is largely valid for RC∕RL ≤ 0.01 and (S02)2 ≥ 0.8, τe MF represents the significantly reduced potential-dependent renormalized value of τ. Hence, the application of this formula to calculate NMR variables is appropriate in this parameter range, but associating τe with the local motion correlation time is inappropriate. Means to derive τ from τe are provided. For a cosine squared potential, the cone-model-based MF formula that relates τe to τ can also be used. Deriving τ from τe is important for proper characterization of the site-specific local motion and in the context of τ-dependent MF functionalities. Mode-coupling dominates the extended MF regime where SRLS must be invariably used. Eigenmode and spectral density analysis is provided in this study for the two parameter ranges associated with N−H bond motion.
|
|
|
Mode-Coupling Analysis of 15N CSA-15N-1H Dipolar Cross-Correlation in Proteins. Rhombic Potentials at the N-H Bond
E. Meirovitch, Y.E. Shapiro, V. Tugarinov, Z. Liang, and J.H. Freed.
J. Phys. Chem. B 107, 9883-9897 (2003).
<doi: 10.1021/jp030501h>
Publication #293
|
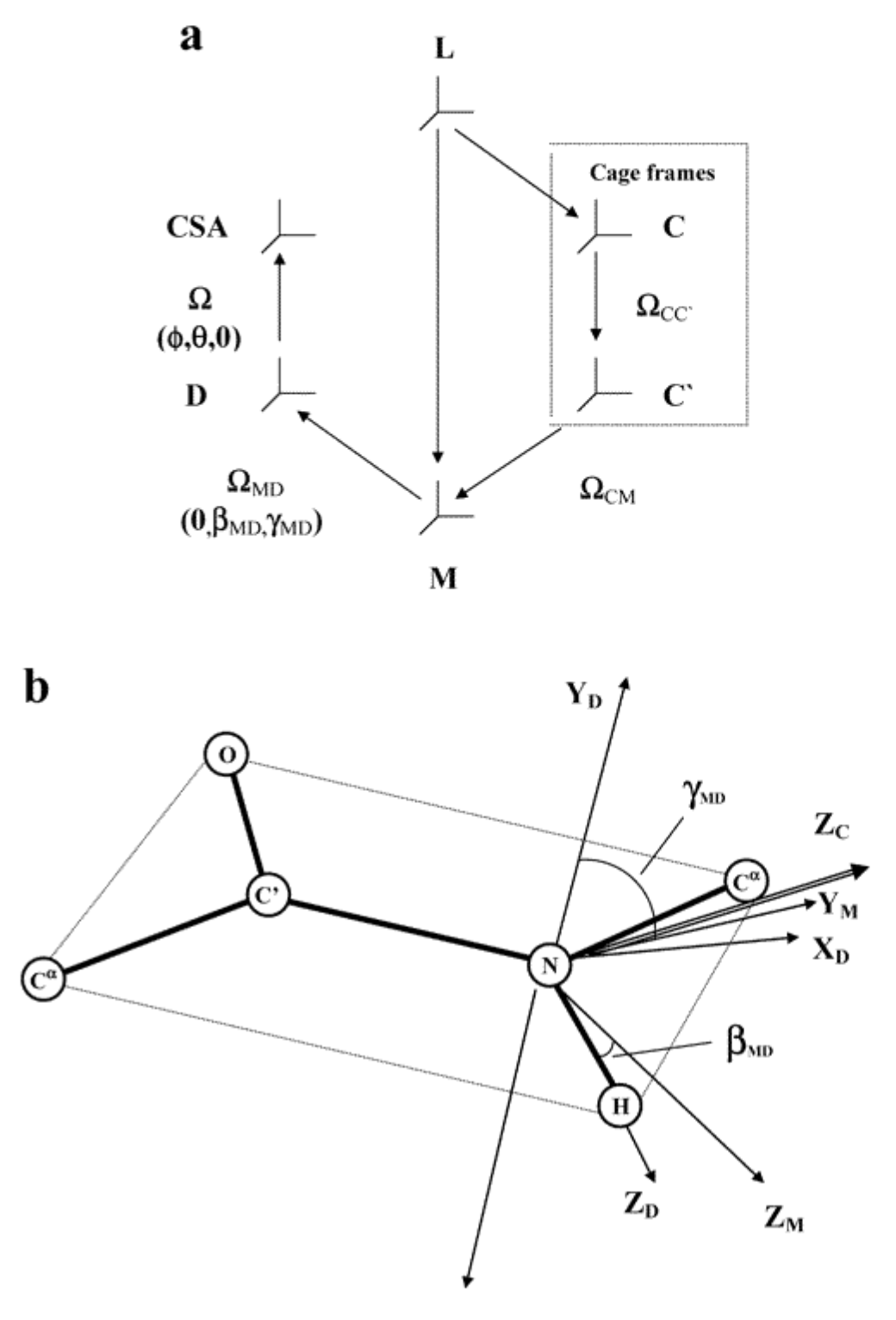
|
|
ABSTRACT: 15N CSA−15N-1H dipolar cross-correlation (ηxy) in proteins has been treated thus far with the model-free (MF) approach, where the global (RC) and local (RL) motions are assumed to be decoupled as a consequence of RL ≫ RC. In the context of ηxy, it is additionally assumed that the local motion is very fast and highly symmetrical. We have recently applied to auto-correlated 15N spin relaxation the slowly relaxing local structure (SRLS) approach, which accounts rigorously for mode-coupling. SRLS can analyze ηxy for arbitrary time scale separations between RL and RC. Simulations of ηxy for slow local motions are presented herein for the first time. Experimental ηxy values of RNase and AKeco could not be reproduced from best-fit parameters generated by data fitting that used axial potentials. Calculations showed they are reproducible using rhombic coupling/ordering potentials. The conformational exchange term, Rex, "absorbs" potential rhombicity when axial potentials are used to fit the data. 15N CSA variability and RC anisotropy are shown to have a small effect on the analysis. The shape of the rhombic potentials detected corresponds to nearly "planar YMXM ordering" (M denotes the local ordering frame). This potential form is consistent with the known geometry of the peptide plane, the SRLS dynamic model in the RL⊥ ≈ RC regime, and makes possible associating the local director with the Ci−1α-Ciα axis. It is shown that back-calculation of NMR variables from the best-fit parameters and comparison with the experimental counterparts is an effective tool for identifying inappropriate elements of the dynamic model and can thereby help improve it.
|
|
|
Hydration, Structure, and Molecular Interactions in the Headgroup Region of Dioleoylphosphatidyl-Choline Bilayers: An Electron Spin Resonance Study
M. Ge and J.H. Freed.
Biophys. J. 85, 4023-40. (2003).
<doi: 10.1016/S0006-3495(03)74816-4>
PMID:
14645091
PMCID:
PMC1303703
Publication #292
|
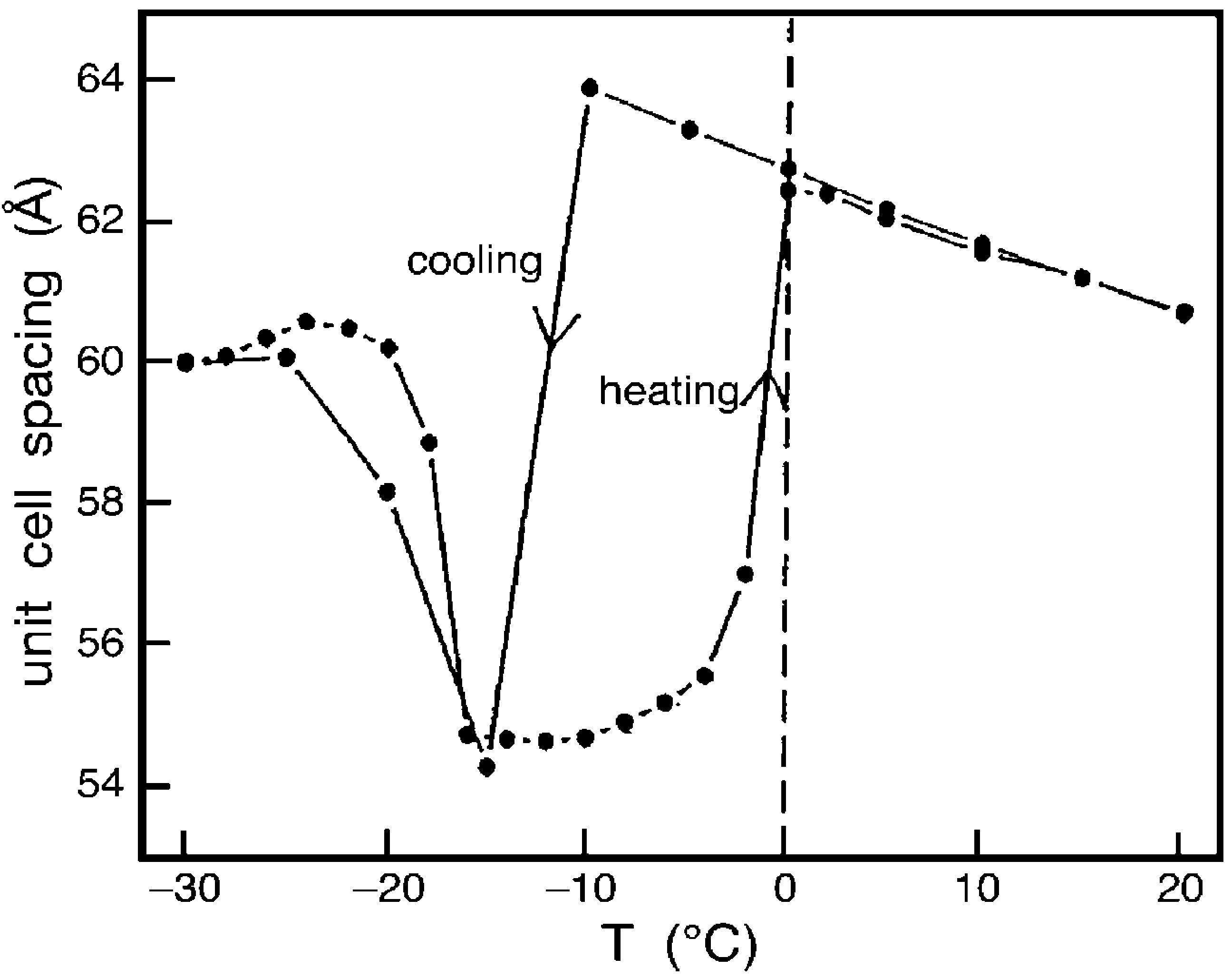
|
|
ABSTRACT: The relationship between bilayer hydration and the dynamic structure of headgroups and interbilayer water in multilamellar vesicles is investigated by electron spin resonance methods. Temperature variations of the order parameter of a headgroup spin label DPP-Tempo in DOPC in excess water and partially dehydrated (10wt % water) show a cusp-like pattern around the main phase transition, Tc. This pattern is similar to those of temperature variations of the quadrupolar splitting of interbilayer D2O in PC and PE bilayers previously measured by 2H NMR, indicating that the ordering of the headgroup and the interbilayer water are correlated. The cusp-like pattern of these and other physical properties around Tc are suggestive of quasicritical fluctuations. Also, an increase (a decrease) in ordering of DPP-Tempo is correlated with water moving out of (into) interbilayer region into (from) the bulk water phase near the freezing point, Tf. Addition of cholesterol lowers Tf, which remains the point of increasing headgroup ordering. Using the small water-soluble spin probe 4-PT, it is shown that the ordering of interbilayer water increases with bilayer dehydration. It is suggested that increased ordering in the interbilayer region, implying a lowering of entropy, will itself lead to further dehydration of the interbilayer region until its lowered pressure resists further flow, i.e., an osmotic phenomenon.
|
|
|
ESR and Molecular Dynamics
J.H. Freed.
In Biomedical EPR, Part B: Methodology, Instrumentation and Dynamics, S.S. Eaton, G.R. Eaton, and L.J. Berliner, Eds.
Biological Magnetic Resonance 24, Chapter 9, pp. 239-268 (2005). (Kluwer, NY).
|
|
|
ESR and Molecular Dynamics
J.H. Freed.
In Biomedical EPR, Part B: Methodology, Instrumentation and Dynamics, S.S. Eaton, G.R. Eaton, and L.J. Berliner, Eds.
Biological Magnetic Resonance 24, Chapter 9, pp. 239-268 (2005). (Kluwer, NY).
<doi: 10.1007/0-306-48533-8_9>
Publication #291
|

|
|
ABSTRACT: The development of ESR for the study of spin-relaxation and molecular dynamics of organic radicals and spin labels in fluids is reviewed from a historical perspective.
|
|
|
Ordered and Disordered Phases Coexist in Plasma Membrane Vesicles of RBL-2H3 Mast Cells. An ESR Study
M. Ge, A. Gidwani, H.A. Brown, D. Holowka, B. Baird, and J.H. Freed.
Biophys. J. 85, 1278-1288 (2003).
<doi: 10.1016/S0006-3495(03)74563-9>
PMID:
12885671
PMCID:
PMC1303245
Publication #290
|
|
|
ABSTRACT: Four chain spin labels and a spin-labeled cholestane were used to study the dynamic structure of plasma membrane vesicles (PMV) prepared from RBL-2H3 mast cells at temperatures ranging from 22°C to 45°C. Analysis shows that the spectra from most labels consist of two components. The abundant spectral components exhibit substantial ordering that is intermediate between that of a liquid-ordered (Lo) phase, and that of a liquid-crystalline (Lc) phase as represented by model membranes. Also, rotational diffusion rates of the spin labels are comparable to those in the Lo phase. In contrast, the ordering for the less abundant components is much lower. These results indicate that a Lo-like region or phase (the abundant component) and an Lc-like region or phase (the less abundant component) coexist in the PMV. In contrast, membranes reconstituted from extracted lipids exhibit the more ordered phase only. This suggests that membrane-associated proteins are important for the coexistence of Lo-like and Lc-like regions in the plasma membrane. In addition, binding of the myristoylated protein, ARF6 to PMV, leads to a new spectral component for a headgroup lipid spin label that indicates the formation of plasma membrane defects by this low molecular weight GTPase.
|
|
|
End-to-End Correlation for a C-12 Hydrocarbon Chain
S. Wagner, A.A. Nevzorov, J.H. Freed, and R.G. Bryant.
J. Magn. Reson. 160, 161-165 (2003)
<doi: 10.1016/S1090-7807(02)00143-X>
PMID:
12615159
Publication #289
|
|
|
ABSTRACT: The 19F nuclear spin-lattice relaxation rate constants were measured as a function of magnetic field strength for 1,12-diaminododecane labeled at one end with a nitroxide radical and at the other with a trifluoromethyl group. The magnetic relaxation dispersion profile (MRD) reports the spectral density function appropriate to the end-to-end correlation function for the doubly labeled molecule. After extrapolation to zero concentration to eliminate the intermolecular relaxation contribution to relaxation, the resulting intramolecular MRD profile was compared with several model approaches. The rotational model for the spectral density functions as included in the Solomon–Bloembergen–Morgan equations does not describe the data well. The earlier model of Freed for nuclear spin relaxation induced by a freely diffusing paramagnetic co-solute is not rigorous for this case because the paramagnet is tethered to the observed nuclear spin and only a restricted space in the immediate vicinity of the nuclear spin is accessible for pseudo-translational diffusion of one end of the molecule with respect to the other. A generalization of the Torrey model for magnetic relaxation by translational diffusion developed by Nevzorov and Freed, which includes the effect of restrictions imposed by the finite length of the chain, describes the experiment within experimental errors. A simple modification of the Hwang–Freed model that does not specifically include the dynamical effects of the finite tether also provides a good approximation to the data when the tether chain is sufficiently long.
|
|
|
Lipid-Gramicidin Interactions: Dynamic Structure of the Boundary Lipid by 2D-ELDOR
A.J. Costa-Filho, R.H. Crepeau, P.P. Borbat, M. Ge, and J. H. Freed.
Biophys. J. 84, 3364-3378 (2003).
<doi: 10.1016/S0006-3495(03)70060-5>
PMID:
12719265
PMCID:
PMC1302896
Publication #288
|
|
|
ABSTRACT: The use of 2D-electron-electron double resonance (2D-ELDOR) for the characterization of the boundary lipid in membrane vesicles of DPPC and gramicidin A′ (GA) is reported. We show that 2D-ELDOR, with its enhanced spectral resolution to dynamic structure as compared with continuous-wave electron spin resonance, provides a reliable and useful way of studying lipid-protein interactions. The 2D-ELDOR spectra of the end-chain spin label 16-PC in DPPC∕GA vesicles is composed of two components, which are assigned to the bulk lipids (with sharp auto peaks and crosspeaks) and to the boundary lipids (with broad auto peaks). Their distinction is clearest for higher temperatures and higher GA concentrations. The quantitative analysis of these spectra shows relatively faster motions and very low ordering for the end chain of the bulk lipids, whereas the boundary lipids show very high "y-ordering" and slower motions. The y-ordering represents a dynamic bending at the end of the boundary lipid acyl chain, which can then coat the GA molecules. These results are consistent with the previous studies by Ge and Freed (1999) using continuous-wave electron spin resonance, thereby supporting their model for GA aggregation and HII phase formation for high GA concentrations. Improved instrumental and simulation methods have been employed.
|
|
|
A 2D-ELDOR Study of the Liquid Ordered Phase in Multilamellar Vesicle Membranes
A.J. Costa-Filho, Y. Shimoyama, and J.H. Freed.
Biophys. J. 84, 2619-2633, (2003).
<doi: 10.1016/S0006-3495(03)75067-X>
PMID:
12668470
PMCID:
PMC1302828
Publication #287
|
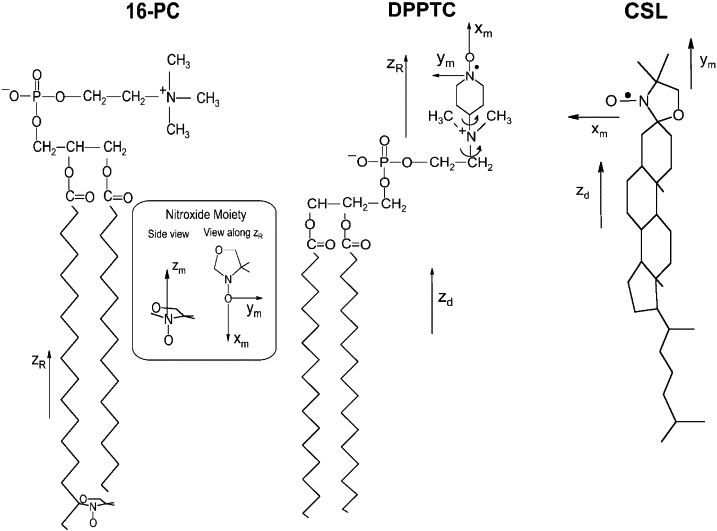
|
|
ABSTRACT: 2D-ELDOR spectroscopy has been employed to study the dynamic structure of the liquid-ordered (Lo) phase versus that of the liquid-crystalline (Lc) phase in multibilayer phospholipid vesicles without (Lc) and with (Lo) cholesterol, using end-chain and headgroup labels and spin-labeled cholestane. The spectra are in most cases found to be dramatically different for these two phases. Thus, visual inspection of the 2D-ELDOR spectra provides a convenient way to distinguish the two phases in membranes. Detailed analysis shows these observations are due to increased ordering in the Lo phase and modified reorientation rates. In the Lo phase, acyl chains undergo a faster rotational diffusion and higher ordering than in the Lc phase, whereas spin-labeled cholestane exhibits slower rotational diffusion and higher ordering. On the other hand, the choline headgroup in the Lo phase exhibits faster motion and reduced but realigned ordering versus the Lc phase. The microscopic translational diffusion rates in the Lo phase are significantly reduced in the presence of cholesterol. These results are compared with previous studies, and a consistent model is provided for interpreting them in terms of the differences in the dynamic structure of the Lo and Lc phases.
|
|
|
Variable-Frequency EPR Study of Mn2+-Doped NH4Cl0.9I0.1 Single Crystal at 9.6, 36, and 249.9 GHz: Structural Phase Transition
S.K. Misra, S.I. Andronenko, G. Rinaldi, P. Chand, K.A. Earle, and J.H. Freed.
J. Magn. Reson. 160, 131-138, (2003).
<doi: 10.1016/S1090-7807(02)00133-7>
PMID:
12615154
Publication #286
|
|
|
ABSTRACT: Multifrequency electron paramagnetic resonance studies on the Mn2+ impurity ion in a mixed single crystal NH4Cl0.9I0.1 were carried out at 9.62 (X-band) in the range 120–295 K, at 35.87 (Q-band) at 77 and 295 K, and at 249.9 GHz (far-infrared band) at 253 K. The high-field EPR spectra at 249.9 GHz are well into the high-field limit leading to a considerable simplification of the spectra and their interpretation. Three magnetically inequivalent, but physically equivalent, Mn2+ ions with their respective magnetic Z-axes oriented along the crystallographic [1 0 0], [0 1 0], [0 0 1] axes were observed. Simultaneous fitting of EPR line positions observed at X-, Q-, and far infra-red bands was performed using a least-squares procedure and matrix diagonalization to estimate accurately the Mn2+ spin-Hamiltonian parameters. The temperature variation of the linewidth and peak-to-peak intensities of the EPR lines indicate the presence of λ-transitions in the mixed NH4Cl0.9I0.1 crystal at 242 and 228 K consistent with those observed in the pure NH4Cl and NH4I crystals, respectively. A superposition-model analysis of the spin-Hamiltonian parameters reveals that the local environment of the Mn2+ ion is considerably reorganized to produce axially symmetric crystal fields about the respective Z-axes of the three magnetically inequivalent ions as a consequence of the vacancy created due to charge-compensation when the divalent Mn2+ ion substitutes for a monovalent NH4+ ion in the NH4Cl0.9I0.1 crystal. This reorganization is almost the same as that observed in NH4Cl and NH4I single crystals, although the latter two are characterized by different, simple cubic and face-centered cubic, structures.
|
|
|
Modern ESR Methods in Studies of the Dynamic Structure of Proteins and Membranes
J.H. Freed.
In EPR in the 2lst. Century , A. Kawamori, J. Yamauchi, and H. Ohta, Eds.
Elsevier Science, Amsterdam, Netherlands, 719-730 (2002).
<doi: 10.1016/B978-044450973-4/50117-7>
Publication #285
|
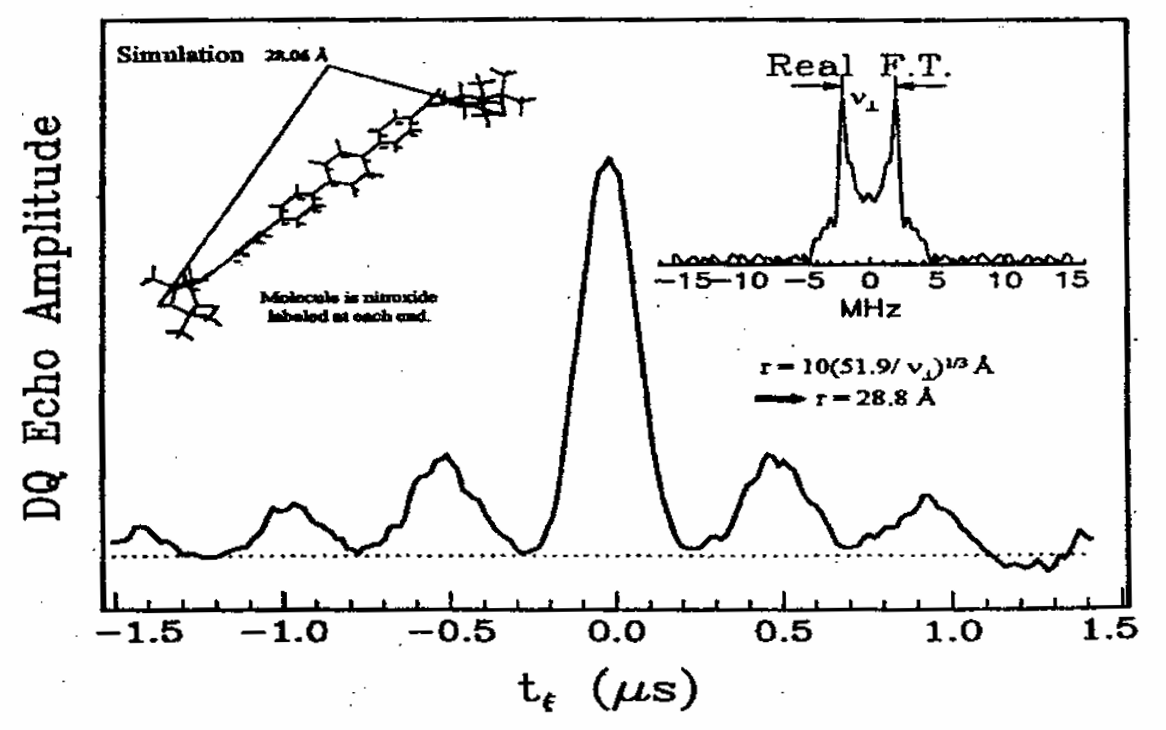
|
|
ABSTRACT: A review of modern electron spin resonance (ESR) techniques for studying the dynamic structure of proteins and membranes using nitroxide spin labels is presented. In particular multiple quantum coherence FT-ESR, multifrequency ESR based on high frequency ESR and two dimensional ESR to study the structure and dynamics of bio-membranes and proteins are illustrated by several examples. Distances in biomolecules have been determined accurately. The details of complex dynamics in proteins and the dynamic structure of membranes were characterized.
|
|
|
Phase Relaxation in a Many-Body System of Diffusing Spins: Slow Motional Limit
A.A. Nevzorov and J.H. Freed.
J. Chem. Phys. 117, 282-287 (2002).
<doi: 10.1063/1.1481764>
Publication #284
|
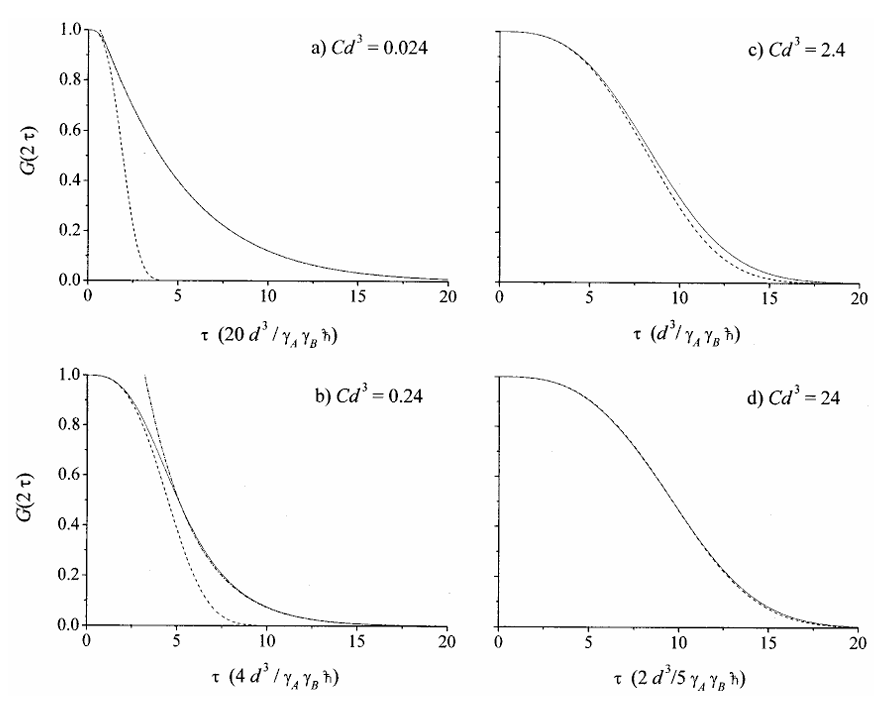
|
|
ABSTRACT: The echo amplitude decay of a diffusing electron-spin-bearing molecule interacting with a diffusing many-spin bath of proton-containing molecules has been studied theoretically in the slow motional limit. Closed-form asymptotic expressions for the short- and long-time behavior of the echo envelope have been obtained. In contrast to the well-studied fast-motional limit, the echo envelope cannot be described by a simple exponential decay, and its rate exhibits a D1∕3T dependence on the relative translational diffusion coefficient, DT For low proton concentrations and long pulse delay times, τ, an exponential τ9∕8-decay of the echo amplitude is found, whereas for high proton concentrations and short τ's the decay exhibits an exponential τ3-behavior. These limiting analytical results are compared with the exact numerical solutions to establish their range of validity.
|
|
|
Domain Flexibility in Ligand-Free and Inhibitor-Bound Escherichia coli Adenylate Kinase Based on a Mode-Coupling Analysis of 15N Spin Relaxation
Y.E. Shapiro, E. Kahana, V. Tugarinov, Z. Liang, J.H. Freed, and E. Meirovitch.
Biochemistry 41, 6271-6281 (2002).
Supporting Information
<doi: 10.1021/bi012132q>
PMID:
12009888
Publication #283
|
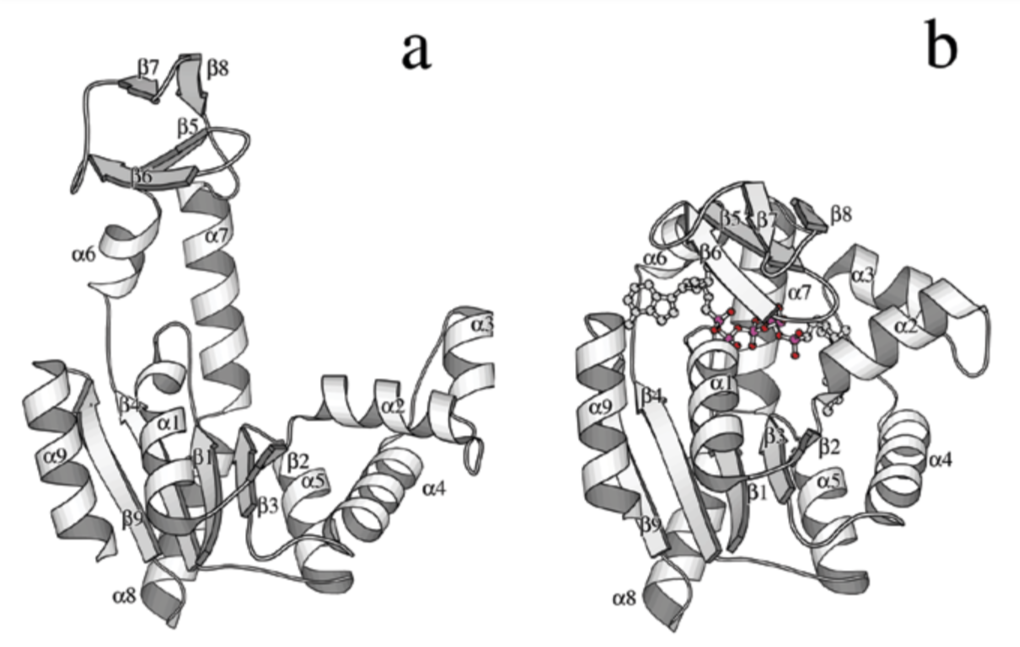
|
|
ABSTRACT: Adenylate kinase from Escherichia coli (AKeco), consisting of a 23.6-kDa polypeptide chain folded into domains CORE, AMPbd, and LID catalyzes the reaction AMP + ATP ↔ 2ADP. The domains AMPbd and LID execute large-amplitude movements during catalysis. Backbone dynamics of ligand-free and AP5A-inhibitor-bound AKeco is studied with slowly relaxing local structure (SRLS) 15N relaxation, an approach particularly suited when the global (τm) and the local (τ) motions are likely to be coupled. For AKeco τm = 15.1 ns, whereas for AKeco*AP5A τm = 11.6 ns. The CORE domain of AKeco features an average squared order parameter, <S2>, of 0.84 and correlation times τf = 5−130 ps. Most of the AKeco*AP5A backbone features <S2> = 0.90 and τf = 33−193 ps. These data are indicative of relative rigidity. Domains AMPbd and LID of AKeco, and loops β1∕α1, α2∕α3, α4∕β3, α5∕β4, and β8∕α7 of AKeco*AP5A, feature a novel type of protein flexibility consisting of nanosecond peptide plane reorientation about the Ci−1α−Ciα axis, with correlation time τ⊥ = 5.6−11.3 ns. The other microdynamic parameters underlying this dynamic model include S2 = 0.13−0.5, τ∥ on the ps time scale, and a diffusion tilt βMD ranging from 12 to 21°. For the ligand-free enzyme the τ⊥ mode was shown to represent segmental domain motion, accompanied by conformational exchange contributions Rex ≤ 4.4 s-1. Loop α4∕β3 and α5∕β4 dynamics in AKeco*AP5A is related to the "energetic counter-balancing of substrate binding" effect apparently driving kinase catalysis. The other flexible AKeco*AP5A loops may relate to domain motion toward product release.
|
|
|
Protein Structure Determination Using Long-Distance Constraints from Double-Quantum Coherence ESR: Study of T4 Lysozyme
P.P. Borbat, H.S. Mchaourab, and J.H. Freed.
J. Am. Chem. Soc. 124, 5304-5314 (2002).
<doi: 10.1021/ja020040y>
PMID:
11996571
Publication #282
|
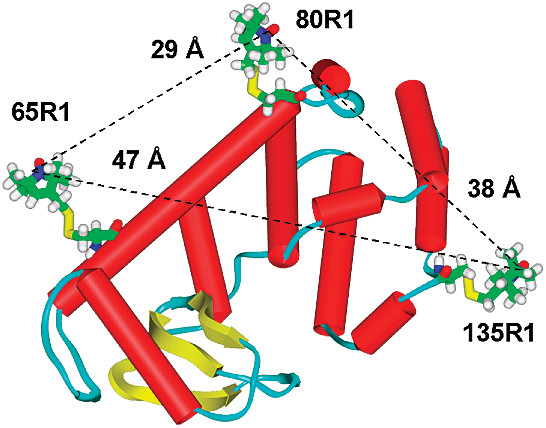
|
|
ABSTRACT: We report the use of a novel pulsed ESR technique for distance measurement, based on the detection of double quantum coherence (DQC), which yields high quality dipolar spectra, to significantly extend the range of measurable distances in proteins using nitroxide spin-labels. Eight T4 lysozyme (T4L) mutants, doubly labeled with methanethiosulfonate spin-label (MTSSL), have been studied using DQC-ESR at 9 and 17 GHz. The distances span the range from 20 Å for the 65∕76 mutant to 47 Å for the 61∕135 mutant. The high quality of the dipolar spectra also allows the determination of the distance distributions, the width of which can be used to set upper and lower bounds in future computational strategy. It is also demonstrated that the shape of these distributions can reveal the presence of multiple conformations of the spin-label, an issue of critical relevance to the structural interpretation of the distances. The distances and distributions found in this study are readily rationalized in terms of the known crystal structure, the characteristic conformers of the nitroxide side chains, and molecular modeling. This study sets the stage for the use of DQC-ESR for determining the tertiary structure of large proteins with just a small number of long-distance constraints.
|
|
|
A Novel View of Domain Flexibility in E. coli Adenylate Kinase Based on Structural Mode-Coupling 15N NMR Relaxation
V. Tugarinov, Y.E. Shapiro, Z. Liang, J.H. Freed and E. Meirovitch.
J. Mol. Biol. 315, 155-170 (2002).
Supporting Information
<doi: 10.1006/jmbi.2001.5231>
PMID:
11779236
Publication #281
|
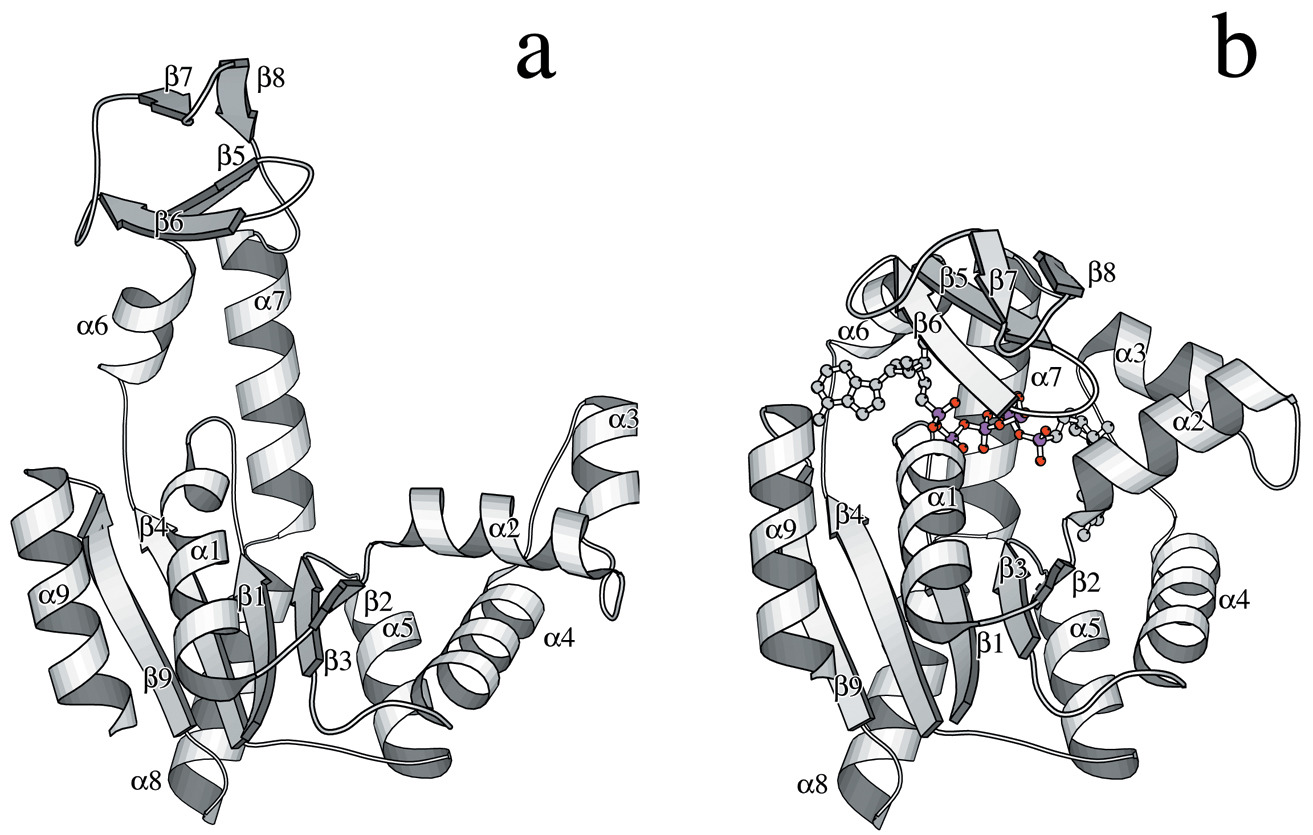
|
|
ABSTRACT: Adenylate kinase from Escherichia coli (AKeco), consisting of a single 23.6 kDa polypeptide chain folded into domains CORE, AMPbd and LID, catalyzes the reaction AMP+ATP→2ADP. In the ligand-free enzyme the domains AMPbd and LID execute large-amplitude movements controlling substrate binding and product release during catalysis. Domain flexibility is investigated herein with the slowly relaxing local structure (SRLS) model for 15N relaxation. SRLS accounts rigorously for coupling between the global and local N-H motions through a local ordering potential exerted by the protein structure at the N-H bond. The latter reorients with respect to its protein surroundings, which reorient on the slower time scale associated with the global protein tumbling. AKeco diffuses globally with correlation time τm=15.1 ns, while locally two different dynamic cases prevail. The domain CORE features ordering about the equilibrium N-H bond orientation with order parameters, S2, of 0.8-0.9 and local motional correlation times, τ, mainly between 5-130 ps. This represents a conventional rigid protein structure with rapid small-amplitude N-H fluctuations. The domains AMPbd and LID feature small parallel (ZM) ordering of S2=0.2-0.5 which can be reinterpreted as high perpendicular (YM) ordering. M denotes the local ordering/local diffusion frame. Local motion about ZM is given by τ∥≈5 ps and local motion of the effective ZM axis about YM by τ⊥=6-11 ns. ZM is tilted at approximately 20° from the N-H bond. The orientation of the YM axis may be considered parallel to the Ci−1α-Ciα axis. The τ⊥ mode reflects collective nanosecond peptide-plane motions, interpretable as domain motion. A powerful new model of protein flexibility∕domain motion has been established. Conformational exchange (Rex) processes accompany the τ⊥ mode. The SRLS analysis is compared with the conventional model-free analysis.
|
|
|
Single-Crystal EPR Studies of Transition-Metal Ions in Inorganic Crystals at Very High Frequency
S.K. Misra, S.I. Andronenko, K.A. Earle, J.H. Freed.
Appl. Magn. Reson. 21, 549-561 (2001).
<doi: 10.1007/BF03162428>
Publication #280
|
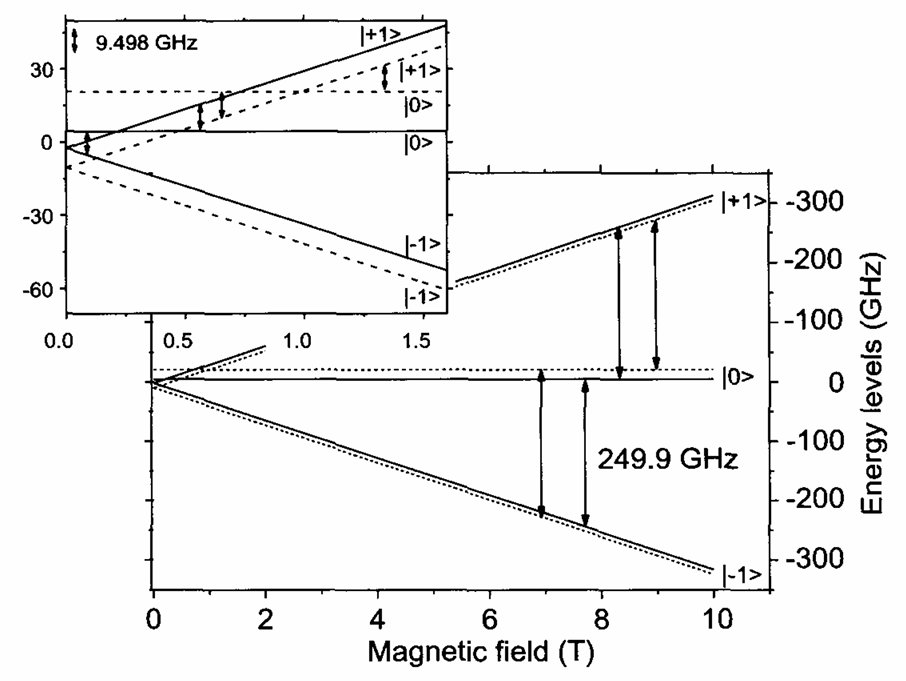
|
|
ABSTRACT: Electron paramagnetic resonance (EPR) single-crystal rotation studies at very high frequency (249.9 GHz) of transition metal ions with electron spins greater than one-half are reported. At 249.9 GHz, the spectra are in the high-field limit despite large zero-field splittings. This leads to a considerable simplification of the spectra, and aids in their interpretation. Single-crystal 249.9 GHz EPR spectra of Ni2+ in Ni2CdCl6 ⋅ 12H2O, Mn2+ (0.2%) in ZnV2O7, and Fe3+ (2%) in CaYAl04 were recorded at 253 K in an external magnetic field of up to 9.2 T, along with those at X-band and Q-band frequencies at 295 K and lower temperatures. The goniometer used at 249.9 GHz for single-crystal rotation is based on a quasi-optical design and is an integral part of a special Fabry-Pérot resonator. The values of the spin-Hamiltonian parameters were estimated from a simultaneous fitting of all of the observed line positions at several microwave frequencies recorded at various orientations of each crystal with respect to the external magnetic field with least-squares fitting in conjunction with matrix diagonalization. Estimates of zero-field splitting parameter D at room temperature are: for Ni2+, about −31 GHz (site I) and about −7 GHz (site II); for Mn2+, about 6 GHz; and for Fe3+, about 29 GHz.
|
|
|
A Multifrequency ESR Study of the Complex Dynamics of Membranes
Y. Lou, M. Ge, and J.H. Freed.
J. Phys. Chem. B 105, 11053-11056 (2001).
<doi: 10.1021/jp013226c>
Publication #279
|
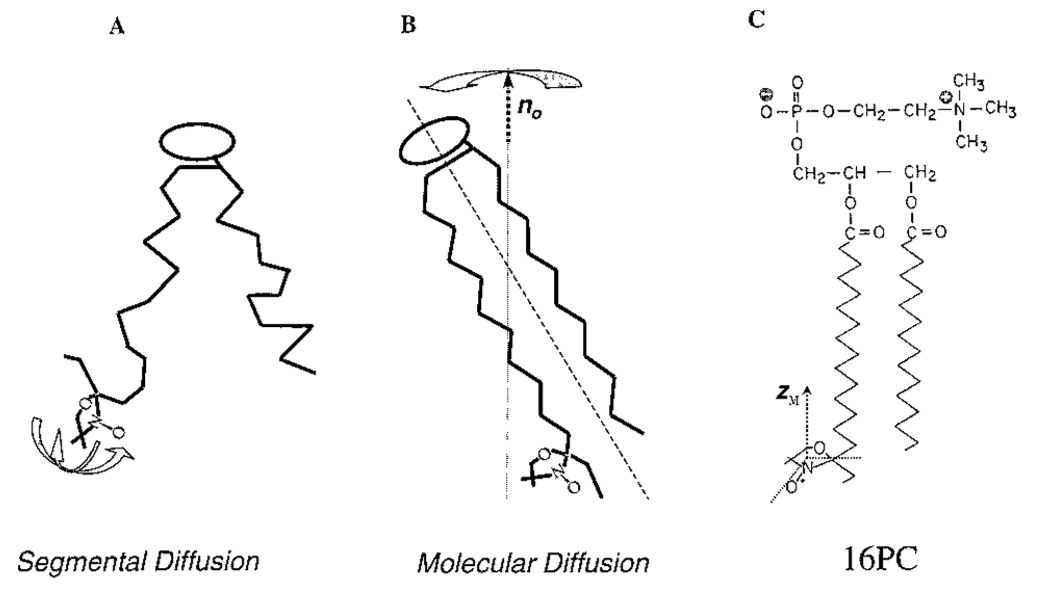
|
|
ABSTRACT: A combined 250 and 9 GHz ESR study was performed on membrane vesicles composed of pure lipid (DPPC) and of DPPC:cholesterol in a 1:1 molar ratio using the end chain labeled lipid, 16-PC. It is shown that the 250 GHz spectra represent a "fast time-scale" such that the overall restricted motion of the lipid in the membrane is frozen out, but it is sensitive to the internal dynamics of the end chain. The 9 GHz spectra are, however, sensitive to the overall motion as well. This combined study thus permitted the separation of both types of motion. It is shown that the addition of cholesterol has a large effect on the end chain dynamics by restricting its motion while increasing motional rates, whereas its effects on the overall lipid motion are modest. This leads to a clearer characterization of the dynamic structure of the cholesterol-rich liquid-ordered phase as compared to the liquid crystalline phase. This study thus shows the significant potential of a multifrequency ESR approach to the complex dynamics of membranes.
|
|
|
A Many-Body Analysis of the Effects of the Matrix Protons and their Diffusional Motion on Electron Spin Resonance Line Shapes and Electron Spin Echoes
A.A. Nevzorov and J.H. Freed.
J. Chem. Phys. 115, 2416-2429 (2001)
<doi: 10.1063/1.1382817>
Publication #278
|
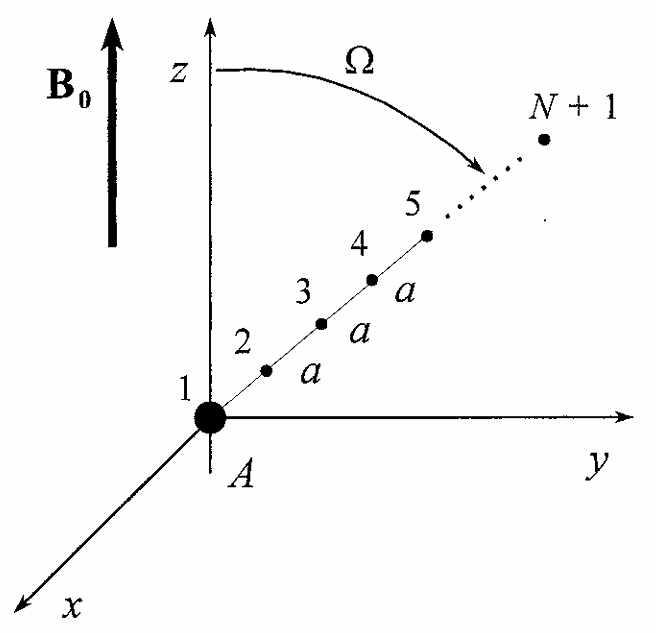
|
|
ABSTRACT: The method for treating the evolution of the density matrix developed in the accompanying paper for many-spin systems is applied here for calculating magnetic resonance signals of a spin A interacting with a bath of N identical spins B. Spins B are assumed to have much smaller gyromagnetic ratios than the spin A (e.g., the former are nuclear spins, I and the latter is an electron spin, S). The experimentally observed quadratic dependence of the spin-echo envelope decay on concentration and time is explained from considering the dipolar coupling of spin A to all the B spins in the presence of B–B dipolar interactions. It is shown that the spin-echo envelope decay in the rigid limit is due to the interaction of the A spin with the coherent many-body states of the coupled spins B via the nuclear flip-flop terms I±I∓ which becomes a dissipative mechanism in the thermodynamic limit. This represents a more rigorous analysis than simplified models based on an incoherent version of "spin diffusion," and it leads to good quantitative agreement with experiment. Moreover, this analysis represents a unified description of both the modulation and decay of the A-spin echoes. Spin echoes and line shapes for the A–BN systems are also calculated for finite motions which randomize the B spins. Even for very slow motions (modeled as translational diffusion) an effective mechanism for spin-echo envelope decay is generated, which readily overtakes the coherent mechanism in importance. The intensity distribution for the forbidden components in the A-spin line shape resulting from multiquantum transitions of the B spins caused by the pseudosecular interaction terms SzI± is calculated. In the rigid limit it is found to behave like a Poisson distribution.
|
|
|
Direct-Product Formalism for Calculating Magnetic Resonance Signals in Many-Body Systems of Interacting Spins
A.A. Nevzorov and J.H. Freed.
J. Chem. Phys. 115, 2401-2415 (2001).
<doi: 10.1063/1.1382816>
Publication #277
|
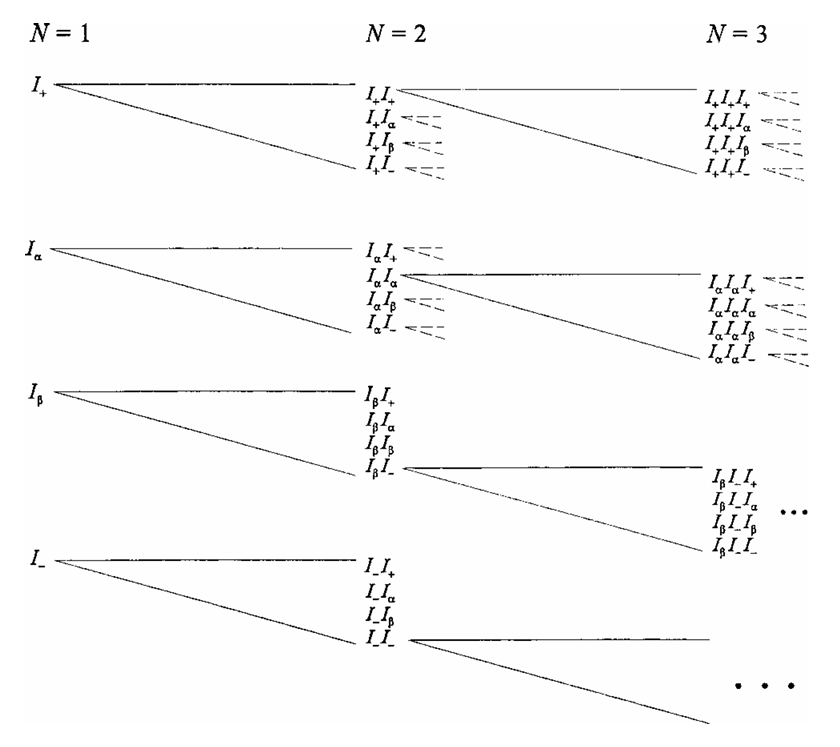
|
|
ABSTRACT: Modern applications of nuclear magnetic resonance and electron spin resonance, and especially quantum computing problems call for more effective formalisms to describe relaxation and evolution of various orders of coherence in the presence of motions of the interacting spins. Here we suggest a formulation for the description of multiquantum spin states based on direct-product structures that take into account the inherent permutation symmetries and quantum coherences of a multispin system. Convenient recursion relations are obtained for the matrix representations of the N-body Hamiltonian superoperator and pulse propagators. This allows one to obtain compact expressions for the evolution of magnetic resonance signals when the dipolar interactions amongst spin-bearing molecules are modulated by their motions. These expressions include the free-induction decay and solid echoes, as well as the decay of higher-order coherences under the assumption of statistical independence of the motions of the spin-bearing molecules. Exact results, not requiring this assumption, are obtained for the case of the truncated dipolar Hamiltonian. Important phenomena that arise in multispin systems, such as instantaneous diffusion and spectral diffusion arising from motions, are studied more rigorously by solving the equation for the time evolution of the spin-density states. The many-body magnetic-resonance signals in the presence of motions are obtained by solving the appropriate stochastic Liouville equations. These solutions may be compared to solid-echo experiments to extract translational diffusion coefficients even in the slow motional regime.
|
|
|
A Structural Mode-Coupling Approach to 15N NMR Relaxation in Proteins
V. Tugarinov, Z. Liang, Y.E. Shapiro, J.H. Freed, and E. Meirovitch.
J. Am. Chem. Soc. 123, 3055-3063 (2001).
Supporting Information
<doi: 10.1021/ja003803v>
PMID:
11457016
Publication #276
|
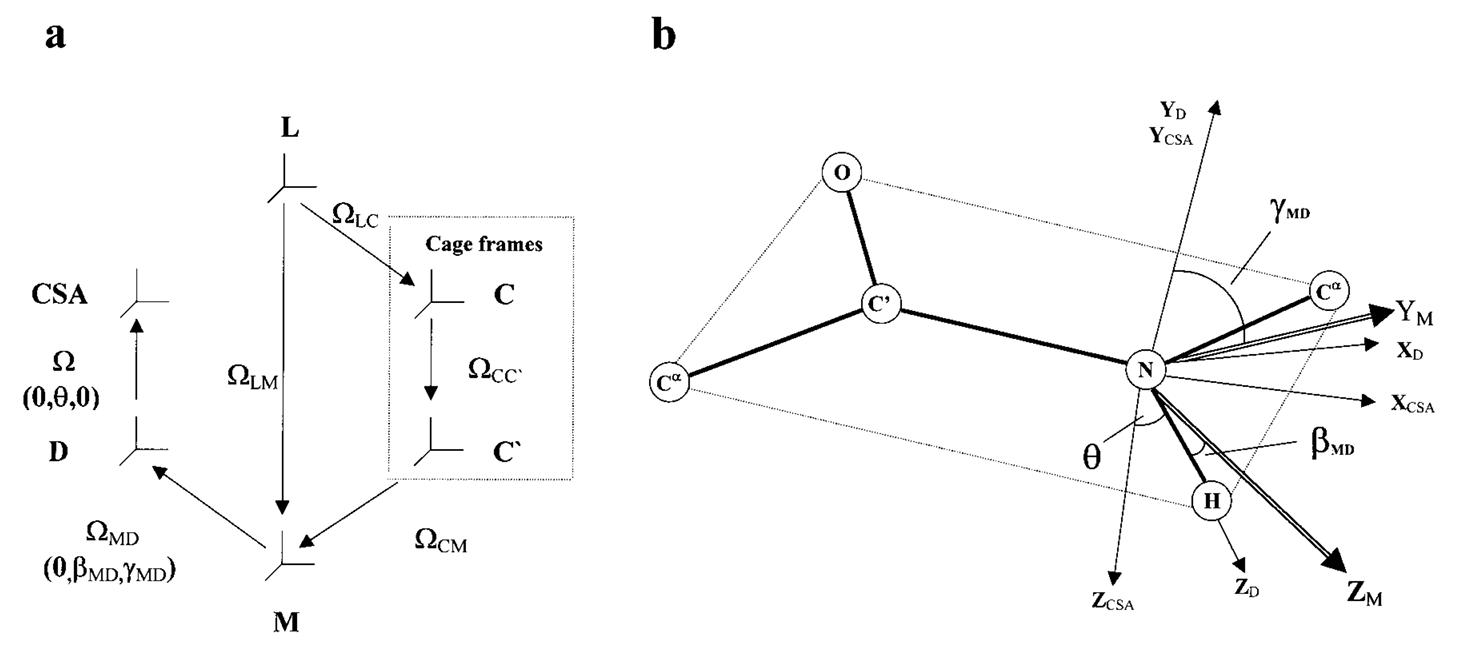
|
|
ABSTRACT: The two-body Slowly Relaxing Local Structure (SRLS) model was applied to 15N NMR spin relaxation in proteins and compared with the commonly used original and extended model-free (MF) approaches. In MF, the dynamic modes are assumed to be decoupled, local ordering at the N−H sites is represented by generalized order parameters, and internal motions are described by effective correlation times. SRLS accounts for dynamical coupling between the global diffusion of the protein and the internal motion of the N−H bond vector. The local ordering associated with the coupling potential and the internal N−H diffusion are tensors with orientations that may be tilted relative to the global diffusion and magnetic frames. SRLS generates spectral density functions that differ from the MF formulas. The MF spectral densities can be regarded as limiting cases of the SRLS spectral density. SRLS-based model-fitting and model-selection schemes similar to the currently used MF-based ones were devised, and a correspondence between analogous SRLS and model-free parameters was established. It was found that experimental NMR data are sensitive to the presence of mixed modes. Our results showed that MF can significantly overestimate order parameters and underestimate local motion correlation times in proteins. The extent of these digressions in the derived microdynamic parameters is estimated in the various parameter ranges, and correlated with the time scale separation between local and global motions. The SRLS-based analysis was tested extensively on 15N relaxation data from several isotropically tumbling proteins. The results of SRLS-based fitting are illustrated with RNase H from E. coli, a protein extensively studied previously with MF.
|
|
|
ADP Ribosylation Factor 6 Binding to Phosphatidylinositol 4,5-Bisphosphate-Containing Vesicles Creates Defects in the Bilayer Structure: An Electron Spin Resonance Study
M. Ge, J.S. Cohen, H.A. Brown, and J.H. Freed.
Biophys. J. 81, 994-1005 (2001).
<doi: 10.1016/S0006-3495(01)75757-8>
PMID:
11463641
PMCID:
PMC1301569
Publication #275
|
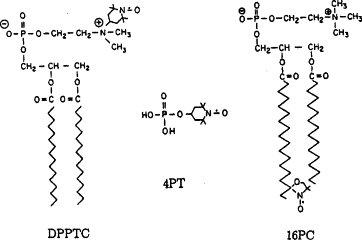
|
|
ABSTRACT: The effects of binding of myristoylated ADP ribosylation factor 6 (myr-ARF6), an activator of phospholipase D (PLD), to a model membrane were investigated using an electron spin resonance (ESR) labeling technique. Initial studies were conducted in vesicles composed of 1-palmitoyl-2-oleoyl phosphatidylethanolamine, dipalmitoylphosphatidylcholine, phosphatidylinositol 4,5-biphosphate (PIP2), and cholesterol. Recombinant ARF6 binding significantly enhances defects in both the headgroup and acyl-chain regions of the membrane, which are revealed by the emergence of sharp components in the spectra from a headgroup label, 1,2-dipalmitoylphosphatidyl-2,2,6,6-tetramethyl-1-piperidinyloxy-choline (DPPTC), and a chain label, 10PC, after myr-ARF6 binding. Binding of non-myristoylated ARF6 (non-ARF6) shows markedly reduced effects. Interestingly, no change in spectra from DPPTC was observed upon myr-ARF6 binding when PIP2 in the vesicles was replaced by other negatively charged lipids, including phosphatidylinositol, phosphatidylserine, and phosphatidylglycerol, even when normalized for charge. The production of the sharp peak appears to be a specific event, because another GTP binding protein, CDC42, which binds PIP2 and activates PLD, fails to induce changes in vesicle structure. These results suggest a previously unappreciated role for ARF in mediating a protein/lipid interaction that produces defects in lipid bilayers. This function may serve as an initial event in destabilizing membrane structure for subsequent membrane fusion or biogenesis of vesicles.
|
|
|
Electron Spin Resonance in Studies of Membranes and Proteins
P.P. Borbat, A.J. Costa-Filho, K.A. Earle, J.K. Moscicki, J.H. Freed.
Science 291, 266-269, (2001).
<doi: 10.1126/science.291.5502.266>
PMID:
11253218
Publication #274
|
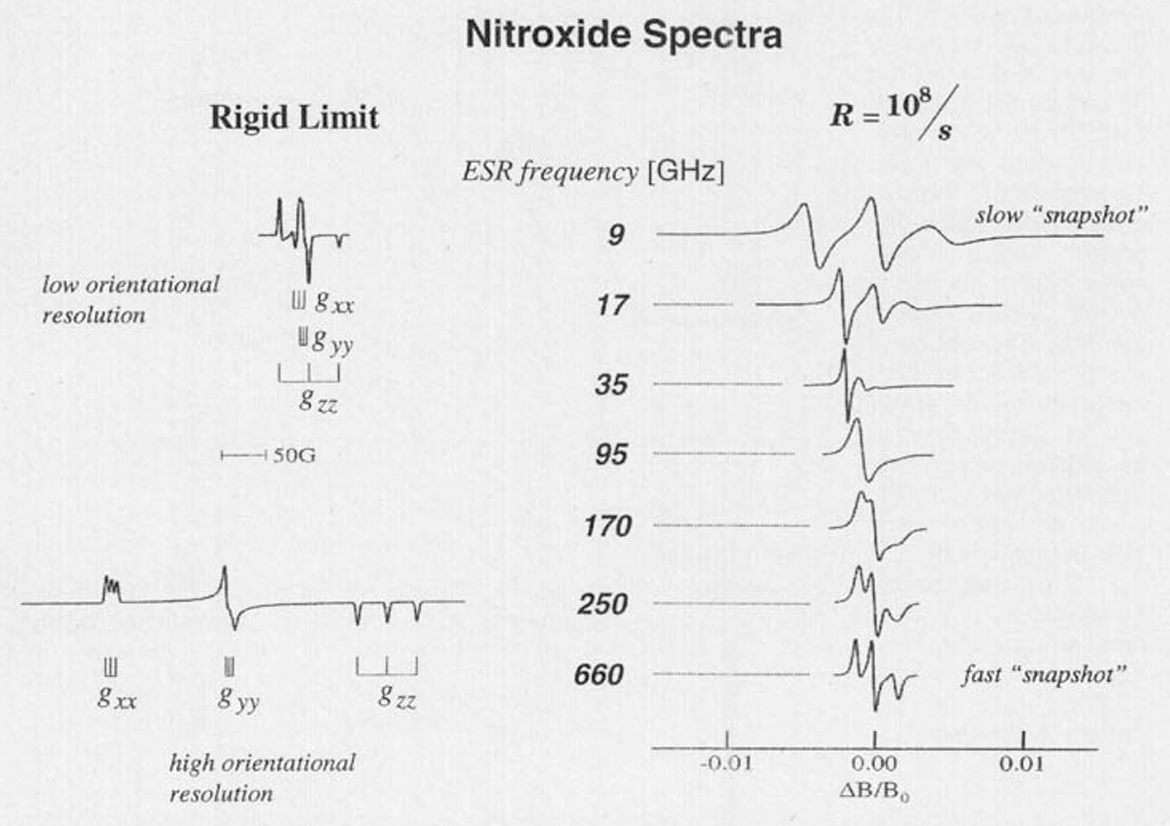
|
|
ABSTRACT: We provide a review of current electron spin resonance (ESR) techniques for studying basic molecular mechanisms in membranes and proteins by using nitroxide spin labels. In particular, nitroxide spin label studies with high-field/high-frequency ESR and two-dimensional Fourier transform ESR enable one to accurately determine distances in biomolecules, unravel the details of the complex dynamics in proteins, characterize the dynamic structure of membrane domains, and discriminate between bulk lipids and boundary lipids that coat transmembrane peptides or proteins; these studies can also provide time resolution to studies of functional dynamics of proteins. We illustrate these capabilities with recent examples.
|
|
|
Segmental Rotational Diffusion of Spin-Labeled Polystyrene in Dilute Toluene Solution by 9 and 250 GHz ESR
J. Pilar, J. Labský, A. Marek, D.E. Budil, K.A. Earle, and J.H. Freed.
Macromolecules 33, 4438-4444 (2000).
<doi: 10.1021/ma0002242>
Publication #273
|
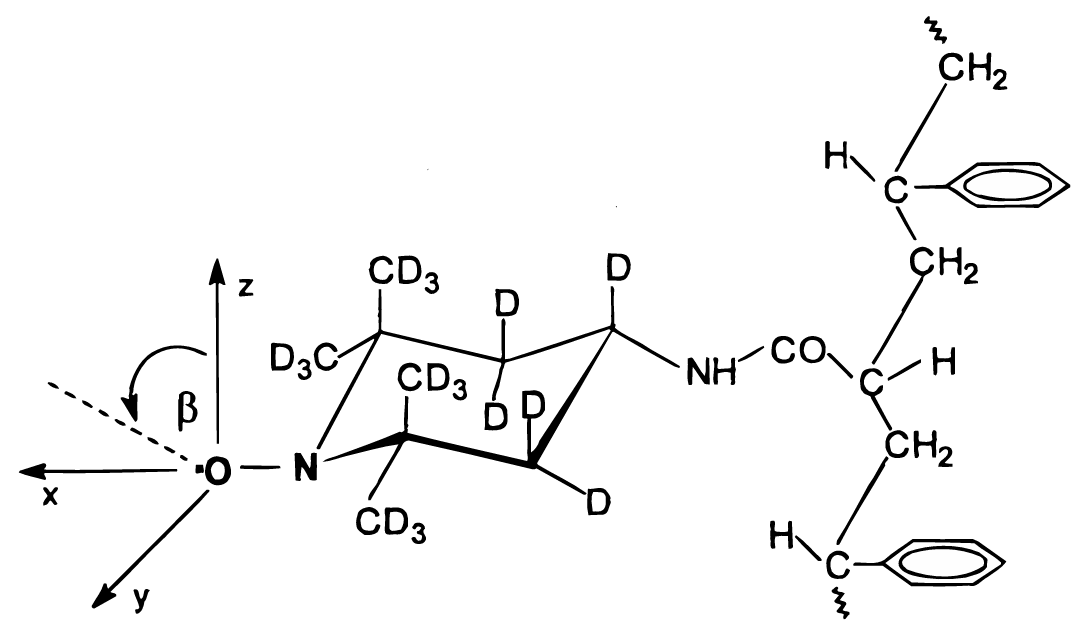
|
|
ABSTRACT: A copolymer of styrene containing less than 5 mol % of chain units spin-labeled by attachment of the nitroxide via a short tether has been synthesized. ESR spectra of dilute toluene solution of the copolymer have been obtained using 9 and 250 GHz ESR. Parameters characterizing polystyrene segmental rotational diffusion in toluene solution over a broad temperature range have been determined from nonlinear least-squares fits of theoretical ESR spectra to the experimental ESR spectra. The model used was that of relatively fast internal rotation of the nitroxide about its tether, with slower polymer chain segmental motion. Together they lead to effective rotational diffusion with an anisotropic diffusion tensor. In addition, constraints in the form of an orientational potential restrict the range of angles over which this diffusion occurs relative to the polymer backbone, and the latter is assumed to reorient on an ultraslow time scale. This is referred to as a model of microscopic order but macroscopic disorder (MOMD). Rates for the slower polymer chain segmental motion (from the 250 GHz spectra) ranged from 3.6 × 108 s-1 at 311 K to 0.15 × 108 s-1 at 215 K, with a substantial orientational potential of about 2kT over this temperature range. Although there was reasonable agreement between the results obtained at 9 and 250 GHz, there were systematic discrepancies such that the orienting potentials obtained from the 250 GHz spectra were about twice (or more) those from the 9 GHz spectra, and the rotational diffusion tensor components from the 250 GHz spectra were at least twice those from the 9 GHz spectra. This implies a breakdown of the MOMD model for the 9 GHz spectra presumably due to their sensitivity to the slower overall tumbling motion at this low spectral frequency. For the faster "time scale" of the 250 GHz spectra, such a slow motion is "frozen out", rendering these spectra consistent with the MOMD model. Nevertheless, the results at both frequencies yielded a common activation energy, Eexp = 20.7 ± 1.5 kJ∕mol, which, when corrected for the viscous flow contribution, yielded an Ea = 11.9 ± 1.5 kJ∕mol, which is in good agreement with recent results from fluorescence studies.
|
|
|
New Technologies in Electron Spin Resonance
J.H. Freed.
Annu. Rev. Phys. Chem. 51, 655-689 (2000).
<doi: 10.1146/annurev.physchem.51.1.655>
PMID:
11031296
Publication #272
|
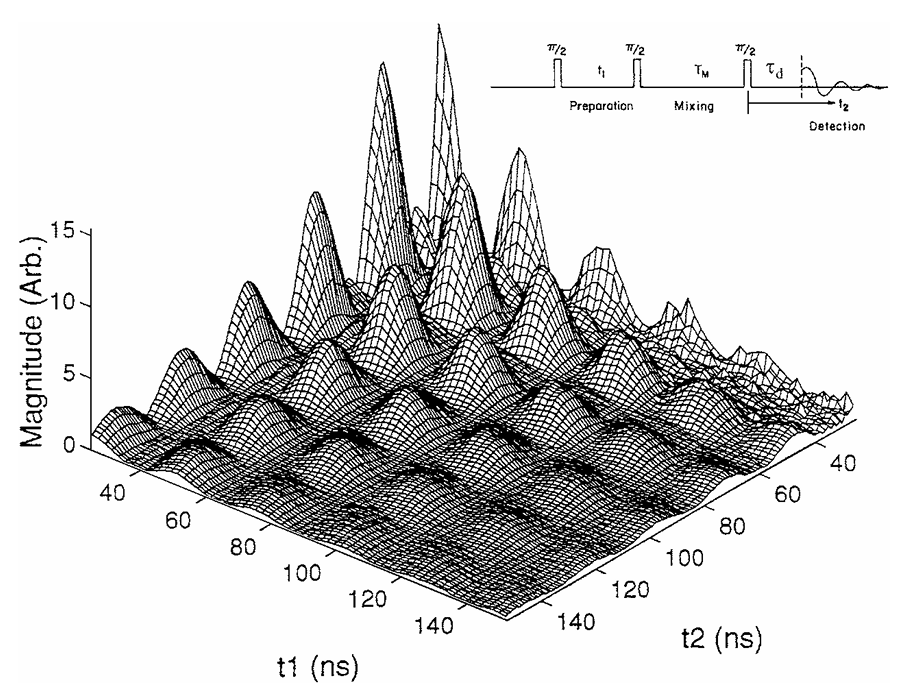
|
|
ABSTRACT: New electron spin resonance (ESR) technologies have been developed, which have led to new and improved applications. (a) The development of two-dimensional Fourier transform (FT) ESR required spectrometers providing intense π∕2 microwave pulses of very short (3–5 ns) duration, wide bandwidths, and very short dead times. It has enabled studies that resolve sophisticated details of molecular dynamics in complex fluids. (b) Methods that produce multiple quantum coherences by pulsed ESR now enable accurate measurements of large distances (>12Å). (c) One of the most important advances has been the extension of ESR to high magnetic fields and high frequencies. This has benefited from the utilization of quasi-optical methods, especially above 150 GHz. The greatly improved orientational resolution and the faster "snapshot" of motions that are provided by ESR at high frequencies enhance studies of molecular dynamics. The use of both high and lower frequencies enables one to unravel faster and slower modes from the complex dynamics of fluids and macromolecules. (d) The development of FT-ESR imaging required substantial pulsed field gradients lasting only 50–100 ns. ESR imaging is effective in studying diffusion in fluids. Areas for further development are also described.
|
|
|
Double Quantum ESR and Distance Measurements
P.P. Borbat and J.H. Freed.
In Distance Measurements in Biological Systems by EPR, L.J. Berliner, S.S. Eaton, and G.R, Eaton, Eds.
Biological Magnetic Resonance 19, Chapter 9, pp. 383-459 (2000). (Kluwer, NY).
|
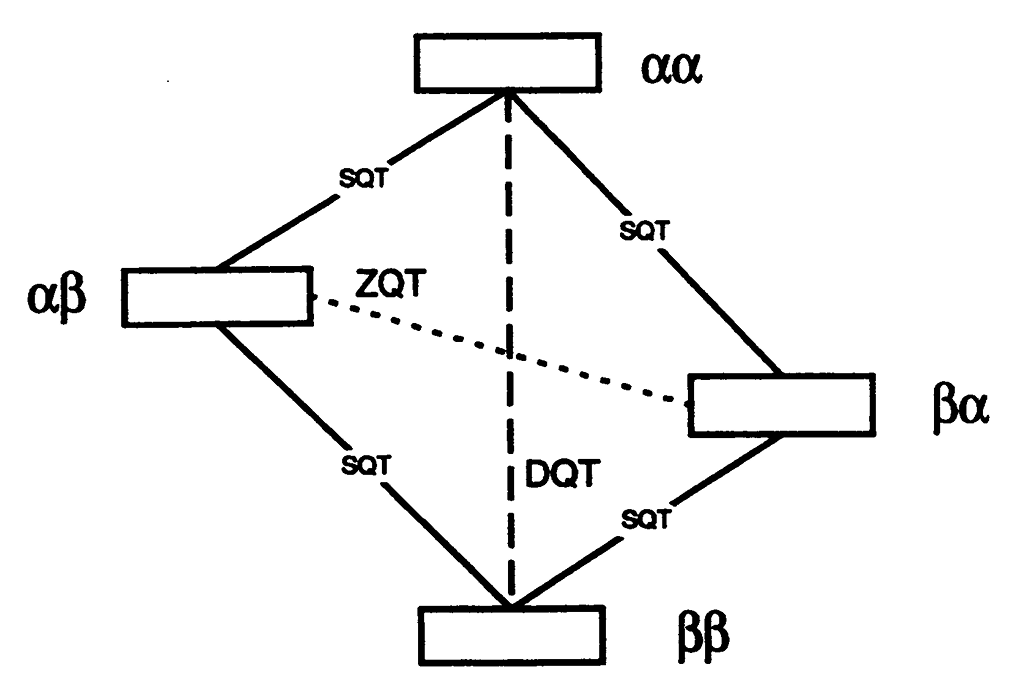
|
|
Double Quantum ESR and Distance Measurements
P.P. Borbat and J.H. Freed.
In Distance Measurements in Biological Systems by EPR, L.J. Berliner, S.S. Eaton, and G.R, Eaton, Eds.
Biological Magnetic Resonance 19, Chapter 9, pp. 383-459 (2000). (Kluwer, NY).
<doi: 10.1007/0-306-47109-4_9>
Publication #271
|

|
|
ABSTRACT: "Allowed" double quantum coherences (DQC) can now be routinely generated in disordered and oriented solids containing nitroxide biradicals and random distributions of stable radicals. The Pake doublets obtained from DQC pathways can be effectively used to determine a broad range of distances in the former case whereas decay constants yield concentrations in the latter. The DQC signals are strong and often comparable to standard single quantum signals. They are free of any large undesirable signals, so the DQ experiment is easy to perform. Their strong intensity permits the study of low concentrations of spins in samples typical of those ordinarily met in the case of doubly-labeled macromolecules such as proteins and polypeptides. The upper range of distances for systems labeled with nitroxides is estimated to be ca. 80 Å. In the limit of non-selective pulses the interpretation of DQC signals becomes independent of complicating geometric features which affect other ESR distance methods. The method is compared to other existing pulse distance measurement techniques and future improvements are also discussed.
|
|
|
An Electron Spin Resonance Study of DNA Dynamics Using the Slowly Relaxing Local Structure Model
Z. Liang, J.H. Freed, R.S. Keyes and A.M. Bobst.
J. Phys. Chem. B 104, 5372-5381 (2000).
<doi: 10.1021/jp994219f>
Publication #270
|
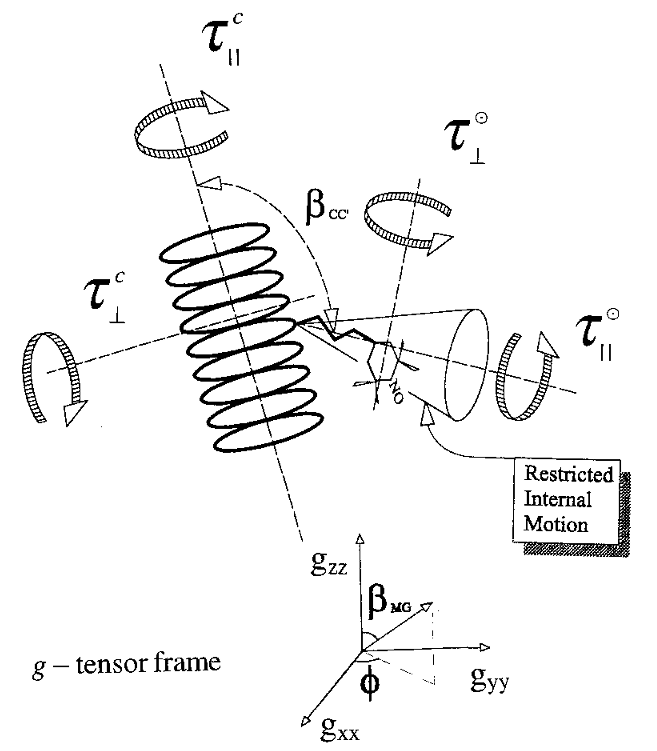
|
|
ABSTRACT: A systematic analysis is presented of the ESR spectra from DNA oligomers of a range of sizes, including a polymer, spin-labeled with nitroxide moieties attached by different tethers. The complexity of the DNA dynamics is dealt with by the use of the general slowly relaxing local structure (SRLS) model, wherein the nitroxide moiety is reorienting in a restricted local environment, which itself is relaxing on a longer time scale. The slower motion describes the global tumbling of the DNA lattice, and the faster motion, the internal dynamics. In the present analysis, the correlation times for the axially symmetric global tumbling were those obtained from hydrodynamic theory, while the correlation times for the internal dynamics and the order parameter, which directly measures its restricted range of motion, were determined by nonlinear least-squares fits to the spectra. The principal result is the observation and characterization of two types of spectra from these labeled DNA systems. These two spectra represent components that differ from each other with respect to their local environments, one a highly restricted site yielding a large order parameter (0.61) and slower internal motions and the other a much less restricted site (with order parameter of 0.18) and faster internal motions. Whereas the one-atom tethered DUTA exhibits both sites (in roughly 9:1 ratio with the more restricted site more prevalent), the two-atom tethered DUMTA and five-atom tethered DUAT exhibit just the more restricted site, but the five-atom tethered DUAP exhibits only the less restricted site. (DUAP differs from DUAT and the others in having a less flexible tether.) It is suggested that the spin labels trapped in the highly restricted∕slow motional site have a stronger interaction with the base than those in the other site. In general, the longer the tether, the faster are the correlation times for internal dynamics. For all tethers, it is found that the correlation time for internal motion perpendicular to the internal symmetry axis systematically becomes slower as the size of the oligomer increases. It is suggested that this may be a manifestation of collective modes of motion of the DNA. It is pointed out that the simpler models used in previous ESR studies are simplified cases of the more realistic SRLS model.
|
|
|
Dipolar Relaxation in a Many-Body System of Spins of 1/2
A.A. Nevzorov and J.H. Freed.
J. Chem. Phys. 112, 1425-1443 (2000).
<doi: 10.1063/1.480709>
Publication #269
|
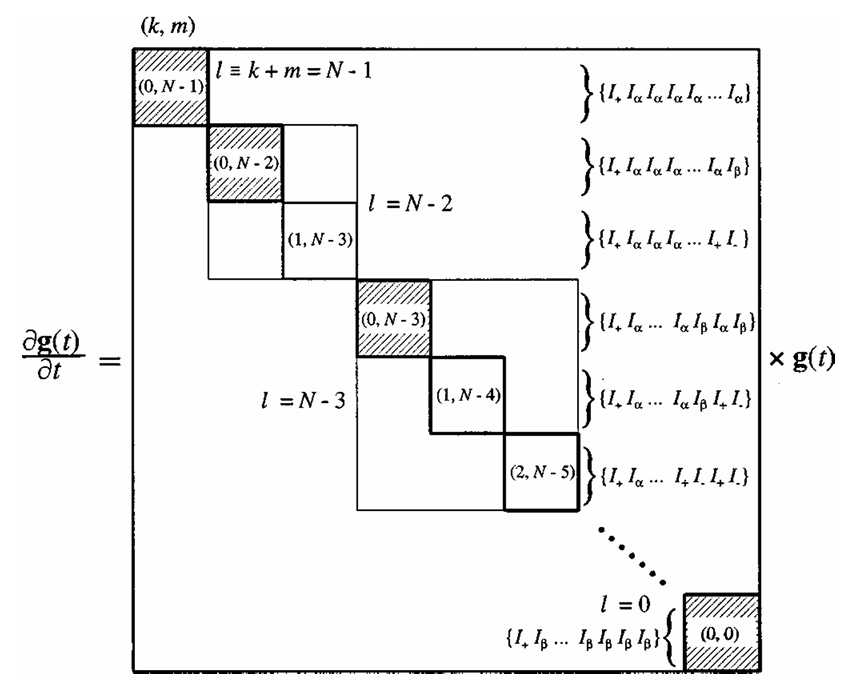
|
|
ABSTRACT: The method utilized in Paper I [J. Chem. Phys. 112, 1413 (2000)] for treating the density matrix equation for a two-spin system in the presence of the dipolar interaction that is randomly modulated by translational diffusion, is extended to a many-body system of identical spins of 1∕2. Generalized cumulant expansions are used, which allow one to take full advantage of the statistical independence of the motions of spins. In the high-temperature approximation (appropriate for dilute solutions), for a single nonselective pulse, the symmetry of the problem allows one to obtain a compact ordered binomial expression for the free-induction decay signal that is related to the two-particle solution, and it still contains the two spin-isochromat components. The latter are evaluated by solving the corresponding stochastic Liouville equation, which allows one to recover in a unified way the two limiting cases including Anderson's result for statistical broadening in a rigid lattice and the classical Torrey–Bloembergen–Redfield expression for the motional narrowing, as corrected by Hwang and Freed. The line shape expression in the thermodynamic limit, i.e., for large numbers of particles in a macroscopic volume, is obtained. It is found that the many-body dipolar line shapes are very close to Lorentzians over the entire motional range studied, with the linewidths proportional to the spin concentration, as predicted earlier for the limiting cases. Linewidths plotted versus the values of the translational diffusion coefficient clearly show the solid-state limit, the motional-narrowing limit, and the intermediate region. The method is extended to describe the behavior of the many-body system in a solid-echo sequence. This enables one to obtain the homogeneous T2's over the whole range of motions. A minimum in T2 is found at approximately the same value of translational diffusion coefficient as was found for the two-spin case in Paper I.
|
|
|
Spin Relaxation by Dipolar Coupling: From Motional Narrowing to the Rigid Limit
A.A. Nevzorov and J.H. Freed.
J. Chem. Phys. 112, 1413-1424 (2000).
<doi: 10.1063/1.480695>
Publication #268
|
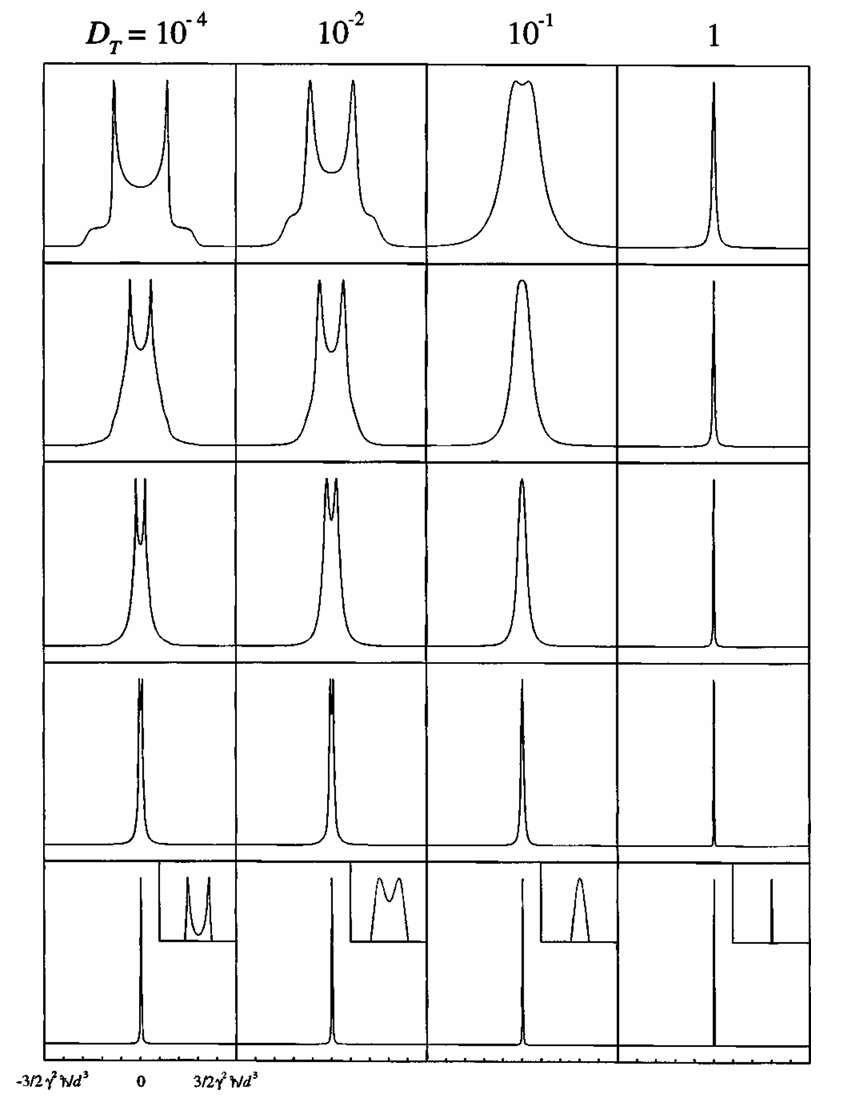
|
|
ABSTRACT: A coupled system of two molecules bearing spins of 1∕2, which are allowed to diffuse relative to each other, is considered. By using a symmetry-adapted basis operator set, the overall density matrix equation is decoupled into two equations for the time-resolved isochromat components, the sum of which yields the observed signal. The appropriate stochastic Liouville equation is solved by a combination of eigenfunction expansion and finite-differences for the angular and radial relative motions, respectively. A full range of spectra from classic Pake patterns in the rigid limit to motionally narrowed Lorentzians is recovered. As an extension of the above approach, the solid-echo experiment is described in terms of the ensemble-averaged isochromats. Homogeneous transverse relaxation times (T2) as a function of the translational diffusion coefficient (DT) are obtained from simulating SECSY (spin-echo correlation spectroscopy) signals, which show distinct T2 minima vs DT. The present method of separating the quantum and stochastic classical variables constitutes a useful approach for treating multiquantum statistical systems, and it can be generalized to an arbitrary number of spins as shown in a companion paper. In the present study we obtained the usual linear dependence of T2 on DT in the motional narrowing (or Redfield) limit, whereas in the slow motional regime a DT−1∕2 dependence is observed, consistent with studies of rotational diffusion. Varying the distance of maximum separation between the two spins (rmax) has virtually no effect on the location of the T2 minimum with respect to DT, implying that the onset of slow motion is essentially independent of rmax.
|
|
|
Multiple-Quantum ESR and Distance Measurements
P.P. Borbat and J.H. Freed.
Chem. Phys. Lett. 313, 145-154 (1999).
<doi: 10.1016/S0009-2614(99)00972-0>
Publication #267
|
|
|
ABSTRACT: It is shown that allowed double-quantum coherences (DQC) can now be routinely generated in disordered and oriented solids containing nitroxide biradicals and random distributions of stable radicals. The Pake doublets obtained from DQC pathways can be effectively used to determine long (˜30 Å) distances in the former case, and concentrations in the latter. The DQC signals are strong and often comparable to standard single-quantum signals. In the limit of non-selective pulses their interpretation becomes independent of complicating features which affect other ESR distance methods.
|
|
|
An Assessment of the Applicability of Multifrequency ESR to Study the Complex Dynamics of Biomolecules
Z. Liang and J.H. Freed.
J. Phys. Chem. B 103, 6384-6396 (1999).
<doi: 10.1021/jp9907746>
Publication #266
|
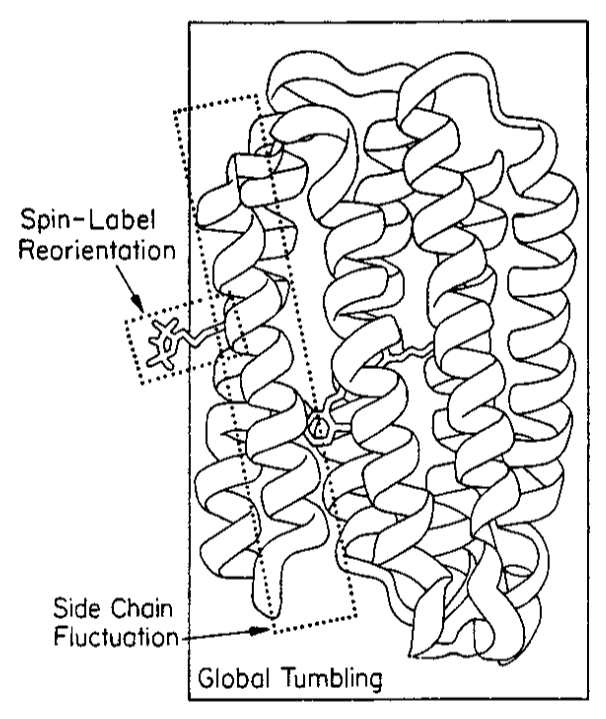
|
|
ABSTRACT: It is shown that the commonly used models for analyzing ESR spectra from nitroxide spin-labeled proteins or DNA systems are special cases of the more general slowly relaxing local structure (SRLS) model, wherein the nitroxide spin probe is taken as reorienting in a restricted local environment, which itself is relaxing on a longer time scale. This faster motion describes the internal dynamics, while the slower motion describes the global tumbling of the macromolecule. By using the SRLS model as the reference, it is shown (1) under what conditions the microscopic-order macroscopic-disorder (MOMD) model, wherein the global tumbling of the macromolecule is in the rigid limit, is valid, and (2) when the fast internal motion (FIM) model, wherein the internal motion is so rapid as to lead to partial averaging of the magnetic tensors, is valid. The frequency dependence of these models is studied. A key general property of high frequency ESR that emerges is that it reports on a faster motional time scale, whereas low frequency ESR reports on a slower motional time scale. It is shown that, in general, the MOMD model is a better approximation for ESR spectra obtained at high frequency (250 GHz), whereas, in general, the FIM model is a better approximation for low frequency (9 GHz) ESR spectra. However, in general, one does not find that the simpler model fits, at a single ESR frequency, to the more complete SRLS model, return correct motional and ordering parameters. The simultaneous fitting of both low and high frequency ESR spectra is thus required to remove such ambiguities and to return all the various dynamic, ordering, and geometric factors that characterize the complex dynamics. This approach is briefly related to recent ESR spectra from the spin-labeled protein, T4 lysozyme, and from spin-labeled DNA nucleosides. In order to better apply the slow-motional SRLS model to macromolecular dynamics, the Polimeno−Freed theory has been extended to the case where the global tumbling is anisotropic and where the angle between the principal axis of the global motion and the preferred orientation of the internal modes of motion is arbitrary.
|
|
|
Electron Spin Resonance Characterization of Liquid Ordered Phase of Detergent Resistant Membranes from RBL-2H3 Cells
M. Ge, K.A. Field, R. Aneja, D. Holowka, B. Baird, and J.H. Freed.
Biophys. J. 77, 925-933 (1999).
<doi: 10.1016/S0006-3495(99)76943-2>
PMID:
10423437
PMCID:
PMC1300383
Publication #265
|

|
|
ABSTRACT: The dynamic structure of detergent-resistant membranes (DRMs) isolated from RBL-2H3 cells was characterized using two different acyl chain spin-labeled phospholipids (5PC and 16PC), a headgroup labeled sphingomyelin (SM) analog (SD-Tempo) and a spin-labeled cholestane (CSL). It was shown, by comparison to dispersions of SM, dipalmitoylphosphatidylcholine (DPPC), and DPPC∕cholesterol of molar ratio 1, that DRM contains a substantial amount of liquid ordered phase: 1) The rotational diffusion rates (R⊥) of 16PC in DRM between −5°C and 45°C are nearly the same as those in molar ratio DPPC∕Chol=1 dispersions, and they are substantially greater than R⊥ in pure DPPC dispersions in the gel phase studied above 20°C; 2) The order parameters (S) of 16PC in DRM at temperatures above 4°C are comparable to those in DPPC∕Chol=1 dispersions, but are greater than those in DPPC dispersions in both the gel and liquid crystalline phases. 3) Similarly, R⊥ for 5PC and CSL in DRM is greater than in pure SM dispersions in the gel phase, and S for these labels in DRM is greater than in the SM dispersions in both the gel and liquid crystalline phases. 4) R⊥ of SD-Tempo in DRM is greater than in dispersions of SM in both gel and liquid phases, consistent with the liquid-like mobility in the acyl chain region in DRM. However, S of SD-Tempo in DRM is substantially less than that of this spin label in SM in gel and liquid crystalline phases (in absolute values), indicating that the headgroup region in DRMs is less ordered than in pure SM. These results support the hypothesis that plasma membranes contain DRM domains with a liquid ordered phase that may coexist with a liquid crystalline phase. There also appears to be a coexisting region in DRMs in which the chain labels 16PC and 5PC are found to cluster. We suggest that other biological membranes containing high concentrations of cholesterol also contain a liquid ordered phase.
|
|
|
Quasioptical Hardware for a Flexible FIR-EPR Spectrometer
K.A. Earle and J.H. Freed.
Appl. Magn. Reson. 16, 247-272 (1999).
<doi: 10.1007/BF03161937>
Publication #264
|
|
|
ABSTRACT: In this paper we present and summarize recent accomplishments of the Freed High Field Electron Paramagnetic Resonance group. In particular, we discuss the application of quasioptical design techniques to instrumentation problems in the far-infrared. We stress that there is no "universal spectrometer" or "universal resonator". Rather, we demonstrate with a variety of examples from the liquid and solid state that the spectrometer configuration and the resonator used should be optimized for the experiment at hand in order to achieve the ultimate in sensitivity. Quasioptical techniques and methods of analysis offer a unified framework to analyze the expected performance of a proposed spectrometer design, as well as suggest the important control parameters for optimizing the sensitivity of a given experiment as we show in the text. The flexibility of quasioptical methods will also be demonstrated with a variety of resonator designs and sample configurations.
|
|
|
Study of Motional Dynamics in Complex Fluids by Very High-Field, Very High-Frequency EPR (VHF-EPR)
A.K. Hassan, J. van Tol, A.L. Maniero, L.C. Brunel, K.A. Earle, and J.H. Freed.
In Physical Phenomena at High Magnetic Fields III, Z. Fisk, L. Gor'kov, J.R. Schrieffer, Eds.
World Scientific NJ, 453-456 (1999)
|
|
|
Study of Motional Dynamics in Complex Fluids by Very High-Field, Very High-Frequency EPR (VHF-EPR)
A.K. Hassan, J. van Tol, A.L. Maniero, L.C. Brunel, K.A. Earle, and J.H. Freed.
In Physical Phenomena at High Magnetic Fields III, Z. Fisk, L. Gor'kov, J.R. Schrieffer, Eds.
World Scientific NJ, 453-456 (1999)
<doi: 10.1142/3945>
Publication #263
|

|
|
ABSTRACT: Very High-field/high-frequency electron paramagnetic resonance (VHF-EPR) has the potential of allowing us to address important questions about fundamental processes in glass-forming systems as the glass transition is approached. We present experimental VHF-EPR spectra taken with a resistive magnet based high field (25 Tesla) EPR spectrometer at different frequencies (525.4275 and 670.463 GHz) over a temperature range from 200 K to 410 K. Two nitroxide spin probes, perdeuterated 2,2, 6,6-tetramethyl-4-methyl aminopiperidinyl-N-oxide (MOTA), and 3,3-dimethyloxazolidinyl-N-oxy-2,3-5a-cholestane (CSL) were used in this VHF-EPR study of the glass former ortho-terphenyl (OTP).
|
|
|
A Multifrequency Electron Spin Resonance Study of T4 Lysozyme Dynamics
J.P. Barnes, Z. Liang, H.S. Mchaourab, J.H. Freed, and Wayne L Hubbell.
Biophys. J. 76, 3298-3306, (1999).
<doi: 10.1016/S0006-3495(99)77482-5>
PMID:
10354455
PMCID:
PMC1300299
Publication #262
|
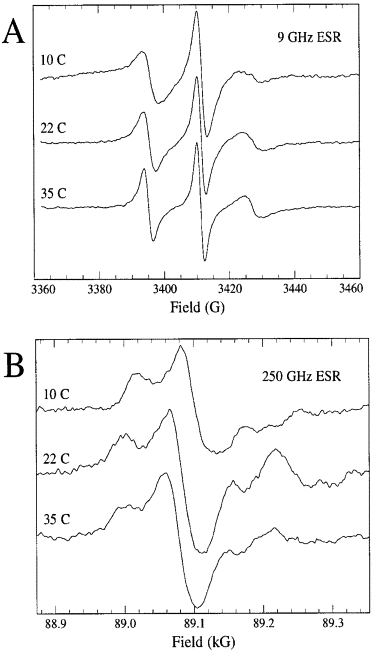
|
|
ABSTRACT: Electron spin resonance (ESR) spectroscopy at 250GHz and 9GHz is utilized to study the dynamics and local structural ordering of a nitroxide-labeled enzyme, T4 lysozyme (EC 3.2.1.17), in aqueous solution from 10°C to 35°C. Two separate derivatives, labeled at sites 44 and 69, were analyzed. The 250-GHz ESR spectra are well described by a microscopic ordering with macroscopic disordering (MOMD) model, which includes the influence of the tether connecting the probe to the protein. In the faster "time scale" of the 250-GHz ESR experiment, the overall rotational diffusion rate of the enzyme is too slow to significantly affect the spectrum, whereas for the 9-GHz ESR spectra, the overall rotational diffusion must be accounted for in the analysis. This is accomplished by using a slowly relaxing local structure model (SRLS) for the dynamics, wherein the tether motion and the overall motion are both included. In this way a simultaneous fit is successfully obtained for both the 250-GHz and 9-GHz ESR spectra. Two distinct motional/ordering modes of the probe are found for both lysozyme derivatives, indicating that the tether exists in two distinct conformations on the ESR time scale. The probe diffuses more rapidly about an axis perpendicular to its tether, which may result from fluctuations of the peptide backbone at the point of attachment of the spin probe.
|
|
|
Multi-Frequency EPR Determination of Zero Field Splitting of High Spin Species in Liquids: Gd(III) Chelates in Water
R.B. Clarkson, A.I. Smirnov, T.I. Smirnova, H. Kang, R.L. Belford, K.A. Earle, and J.H. Freed.
Molec. Phys. 95, 1325-1332 (1998).
<doi: 10.1080/00268979809483262>
Publication #261
|
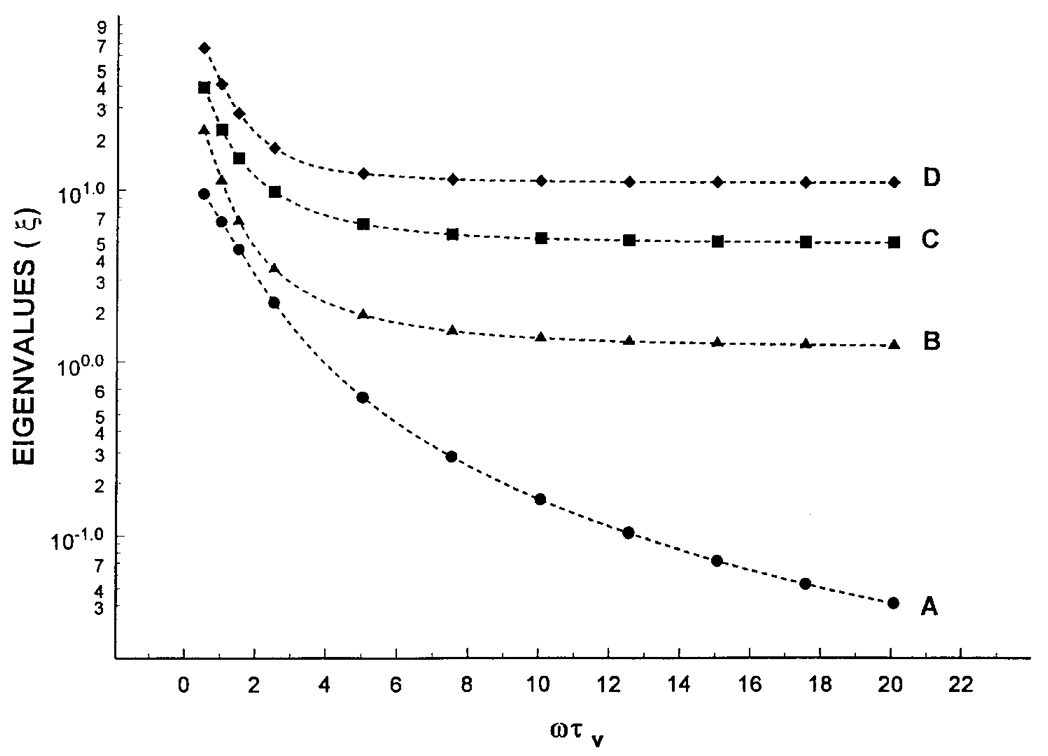
|
|
ABSTRACT: Multi-frequency EPR spectroscopy at 9.5, 35, 94, and 249 GHz has been employed to investigate the zero field splitting (ZFS) of high spin ions in liquids. In particular, experiments are reported on aqueous solutions of DTPA and DOTA chelates of Gd(III), and on the uncomplexed ion, which are relevant to the effectiveness of paramagnetic contrast agents for magnetic resonance imaging (MRI). The field dependence of the centroid of the resonance line, characterized by an effective g factor, geff, has been analysed in order to determine Δ2, the trace of the square of the ZFS matrix. Analysis of the variation in transverse electron spin relaxation (T2e) with experimental frequency provides yet another route to measure Δ2 from EPR data. This analysis also gives τv, a correlation time describing the time-dependent ZFS effect. The ZFS parameters so obtained agree well with results obtained by the analysis of proton nuclear magnetic relaxation dispersion. At 94 GHz, partially resolved spectra from chelated and unchelated Gd(III) were observed. The shifts in resonance field for Gd(III) in these two compounds are due primarily to differences in the magnitude of ZFS. The spectral resolution as a function of frequency exhibits a maximum in the range of our experiments; the resolution disappeared at either higher or lower resonance frequency. Study of ZFS by EPR at multiple high fields offers a new and sensitive route to probe water interactions and chelate dynamics in biologically relevant systems having high spin ions.
|
|
|
Response to "Comment on 'A 250 GHz ESR study of o-terphenyl dynamic cage effects above TC'" [J. Chem. Phys. 109, 10523 (1998)]
K.A. Earle, J.K. Moscicki, A. Polimeno, and J.H. Freed.
J. Chem. Phys. 109 10525-10526 (1998).
<doi: 10.1063/1.477736>
Publication #260
|
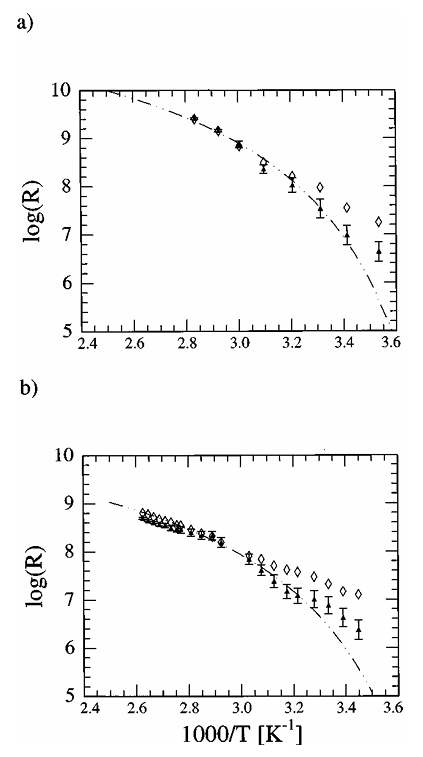
|
|
ABSTRACT: We address the points raised by Giordano and Leporini (GL) and show that accounting properly for the nonexponential decay of the rotational correlation function leads to improved agreement with the Stokes–Einstein–Debye (SED) relation above the crossover temperature TC for those probes 3,3′-dimethyloxazolidinyl-N-oxy-2′,3-5α-cholestane (CSL), and perdeuterated 2,2′,6,6′-tetramethyl-4-methyl aminopiperidinyl-N-oxide) (MOTA) that are well-coupled to the viscous modes of o-terphenyl (OTP) when the average relaxation rate ⅙ ‹τ› is plotted versus 1/T. On the other hand, 2,2′,6,6′-tetramethyl-4-piperidine-N-oxide (PDT) shows simple Arrhenius behavior in this regime, because of weak coupling to the solvent cage, inconsistent with SED, which was clearly shown in our paper. We also suggest that the difference in chemical structure of the PDT probe, studied by us, compared to 2,2′,6,6′-tetramethylpiperidine-N-oxyl (TEMPO), studied by GL, accounts for the difference in the low-temperature relaxation behavior of the two probes.
|
|
|
Electron-Spin Resonance Study of Aggregation of Gramicidin in Dipalmitoylphosphatidylcholine Bilayers and Hydrophobic Mismatch
M. Ge and J.H. Freed.
Biophys. J. 76, 264-280 (1999).
<doi: 10.1016/S0006-3495(99)77195-X>
PMID:
9876140
PMCID:
PMC1302517
Publication #259
|
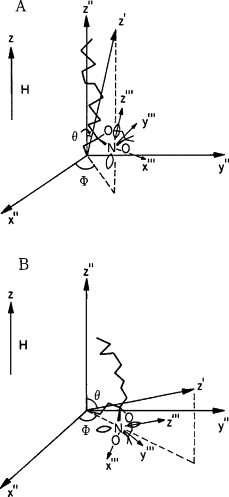
|
|
ABSTRACT: The effect of aggregation of gramicidin A′ (GA) on the phase structure of dipalmitoylphosphatidylcholine (DPPC) multilamellar vesicles was studied by cw-ESR using a chain-labeled lipid (16PC) at temperatures between 30° and 45°C that span the main phase transition of DPPC. Boundary lipids were observed only in dispersions with GA∕DPPC molar ratios >1:15, where GA aggregates. Detailed fits by nonlinear least squares (NLLS) methods are consistent with the boundary lipid being characterized by a large negative order parameter (˜−0.4), indicative of a dynamic bending of the end of the acyl chain, and a substantially reduced motion, about an order of magnitude slower than that of the bulk lipid. The NLLS analysis compares favorably with a recent two-dimensional Fourier transform ESR study on DPPC∕GA vesicles, which accurately discerned the bulk lipid. The detailed ESR observables are discussed in terms of the ordering effect of GA at low concentration of GA, the dissociation of the GA channel and the dynamic bending of the end chain segment of boundary lipid at high concentration of GA, and of HII phase formation induced by GA. It is suggested that these phenomena can be interpreted in terms of the combined effects of partial dehydration of the lipid headgroup by the GA and of the hydrophobic mismatch between GA and DPPC molecules. Substantial hysteresis is observed for heating versus cooling cycles, but only for a GA∕DPPC molar ratio >1:15. This is consistent with the aggregation of GA molecules at high concentrations.
|
|
|
Dynamics and Ordering in Mixed Model Membranes of Dimyristoylphosphatidylcholine and Dimyristoylphosphatidylserine: A 250-GHz Electron Spin Resonance Study Using Cholestane
J.P. Barnes and J.H. Freed.
Biophys. J. 75, 2532-2546 (1998).
<doi: 10.1016/S0006-3495(98)77698-2>
PMID:
9788949
PMCID:
PMC1299928
Publication #258
|
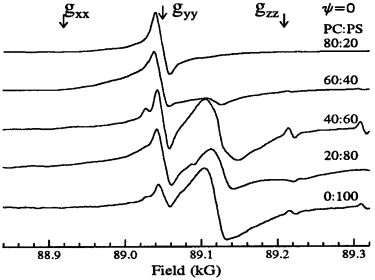
|
|
ABSTRACT: We report here on a 250-GHz electron spin resonance (ESR) study of macroscopically aligned model membranes composed of mixtures of dimyristoylphosphatidylcholine (DMPC) and dimyristoylphosphatidylserine (DMPS), utilizing the nixtroxide-labeled cholesterol analog cholestane (CSL). Two clearly resolved spectral components, distinct in both their ordering and dynamics, are resolved. The major component in membranes composed mostly of DMPC shows typical characteristics, with the long axis of CSL parallel to the bilayer normal with slow (106≤R≤107s−1) rotational diffusion rates, as expected for cholesterol. The second component grows in as the mole fraction of DMPS increases. A detailed analysis shows that CSL senses a local, strongly biaxial environment. Our results imply that the inefficient packing between cholesterol and DMPS occurs probably because of the strong interactions between the PS headgroups, which provide the local biaxiality. Such a packing of the headgroups has been predicted by molecular dynamics simulations but had not been observed experimentally. The analysis of these spectral components was greatly aided by the excellent orientational resolution provided by the 250-GHz spectra. This enabled the key qualitative features of this interpretation to be "read" off the spectra before the detailed analysis.
|
|
|
A "shunt" Fabry–Perot resonator for high-frequency electron spin resonance utilizing a variable coupling scheme
J.P. Barnes and J.H. Freed.
Rev. Sci. Instrum. 69, 3022-3027 (1998).
<doi: 10.1063/1.1149050>
Publication #257
|
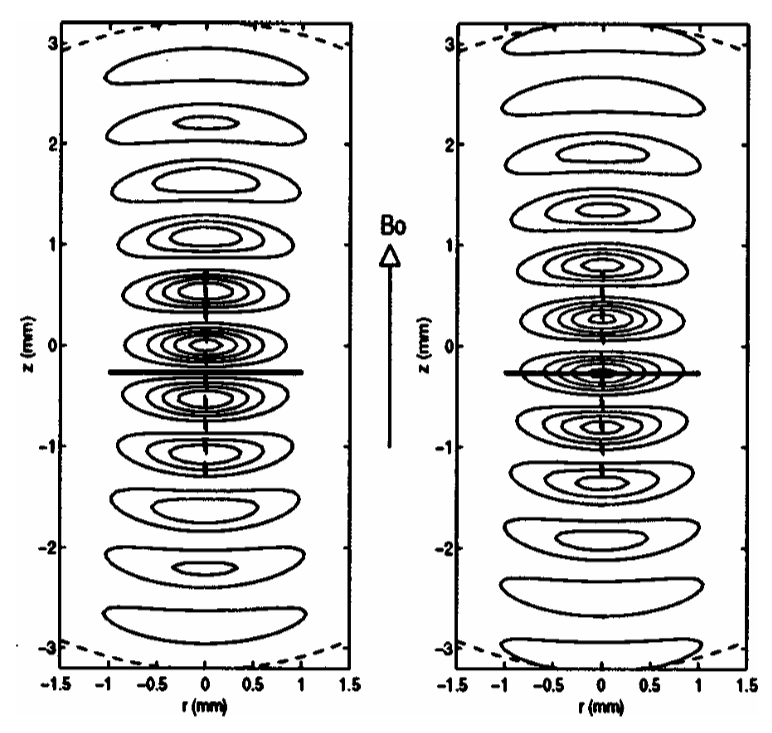
|
|
ABSTRACT: We report on the performance of a Fabry–Perot resonator for far-infrared electron spin resonance (FIR-ESR) at 250 GHz designed to accommodate a thin, disk-shaped sample that must rest with its flat surface perpendicular to the incident FIR beam. This geometry minimizes dielectric losses, making it possible to obtain FIR-ESR spectra of aqueous or lossy samples with a macroscopic ordering, at canonical values of the director tilt of 0° and 90°. The resonator also utilizes an adjustable interferometer to achieve variable coupling in the FIR regime.
|
|
|
Polarity Profiles in Oriented and Dispersed Phosphatidylcholine Bilayers Are Different: An Electron Spin Resonance Study
M. Ge and J.H. Freed.
Biophys. J. 74, 910-917 (1998).
<doi: 10.1016/S0006-3495(98)74014-7>
PMID:
9533702
PMCID:
PMC1302570
Publication #256
|
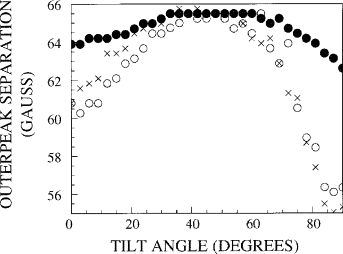
|
|
ABSTRACT: A novel method was utilized to accurately measure the z- component of the nuclear hyperfine interaction tensor, Azz, of a chain-labeled lipid, 16PC, and a headgroup-labeled lipid, dipalmitoylphosphatidyl-tempocholine (DPPTC), in macroscopically oriented dipalmitoylphosphatidylcholine (DPPC) and dimyristoylphosphatidylcholine (DMPC) membranes, which were compared with the Azz values of the two labels in dispersions of the same lipids in the gel phase. We found that the Azz values of 16PC (DPPTC) in the oriented DPPC and DMPC bilayers are ˜1 Gauss smaller (greater) than in the corresponding dispersions. These results indicate that the headgroup region is more polar in macroscopically oriented bilayers than in dispersions, whereas in the chain region, the order in polarity is reversed. This is consistent with previous results on partial molar volumes in the liquid-crystal phase. Differences in the morphology of the macroscopically oriented and dispersed bilayers, which might be responsible, are discussed. Nonlinear least-squares fits of the electron spin resonance spectra of DPPTC in DPPC show that there is a substantial orienting potential in the headgroup region of dispersions that is lipid phase dependent. However, in oriented membrane samples hydrated in 100% relative humidity, this orienting potential is very weak.
|
|
|
Two-Dimensional Electron Spin Resonance and Slow Motions
S. Saxena and J.H. Freed.
J. Phys. Chem. A 101, 7998-8008 (1997).
<doi: 10.1021/jp9717047>
Publication #255
|
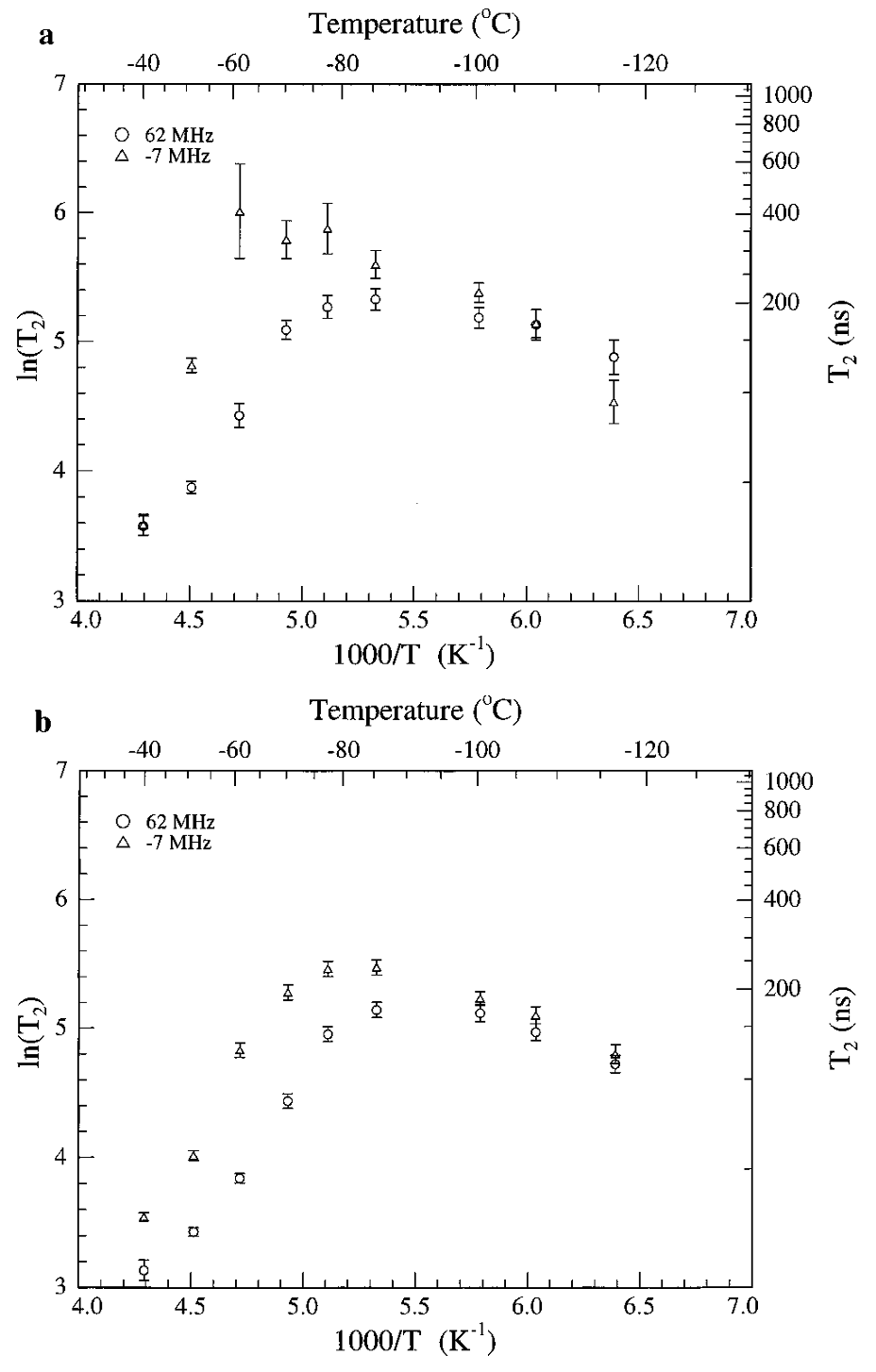
|
|
ABSTRACT: The slow rotational dynamics of a polyproline peptide with a nitroxide labeled at one end in a glassy medium is probed using two-dimensional (2D) electron spin resonance (ESR). The contributions to the homogeneous relaxation time, T2, from the overall and∕or the internal rotations of the nitroxide is elucidated from the COSY spectra. The use of pure absorption spectra allows the variation of T2 across the spectrum to be monitored. It is shown from simulations that the model of anisotropic Brownian diffusion provides semiquantitative agreement with such a variation. In the 2D ELDOR experiment several mechanisms can lead to spectral diffusion, which yields a broadening of the hyperfine (hf) auto-peaks with mixing time. We call these spectral diffusion (SD) cross-peaks. It is shown that at higher temperatures the principal mechanism for the formation of SD cross-peaks is the slow reorientation of the molecule, which modulates the 14N hf and g tensor interactions. A procedure is shown for extracting a correlation time, τc, by monitoring this growth of SD cross-peaks, which is in good agreement with theory. An anomalous temperature dependence of the experimental τc, at very low temperatures, is tentatively attributed to the fast internal rotations of the methyl groups on the nitroxide, which leads to spin-flips of the protons on these methyl groups. The use of pure absorption spectra in 2D ELDOR enhances the sensitivity to these cross-peaks.
|
|
|
Multifrequency Two-Dimensional Fourier Transform ESR: An X/Ku-Band Spectrometer
P.P. Borbat, R.H. Crepeau, and J.H. Freed.
J. Magn. Reson. 127, 155-167 (1997).
<doi: 10.1006/jmre.1997.1201>
PMID:
9281479
Publication #254
|
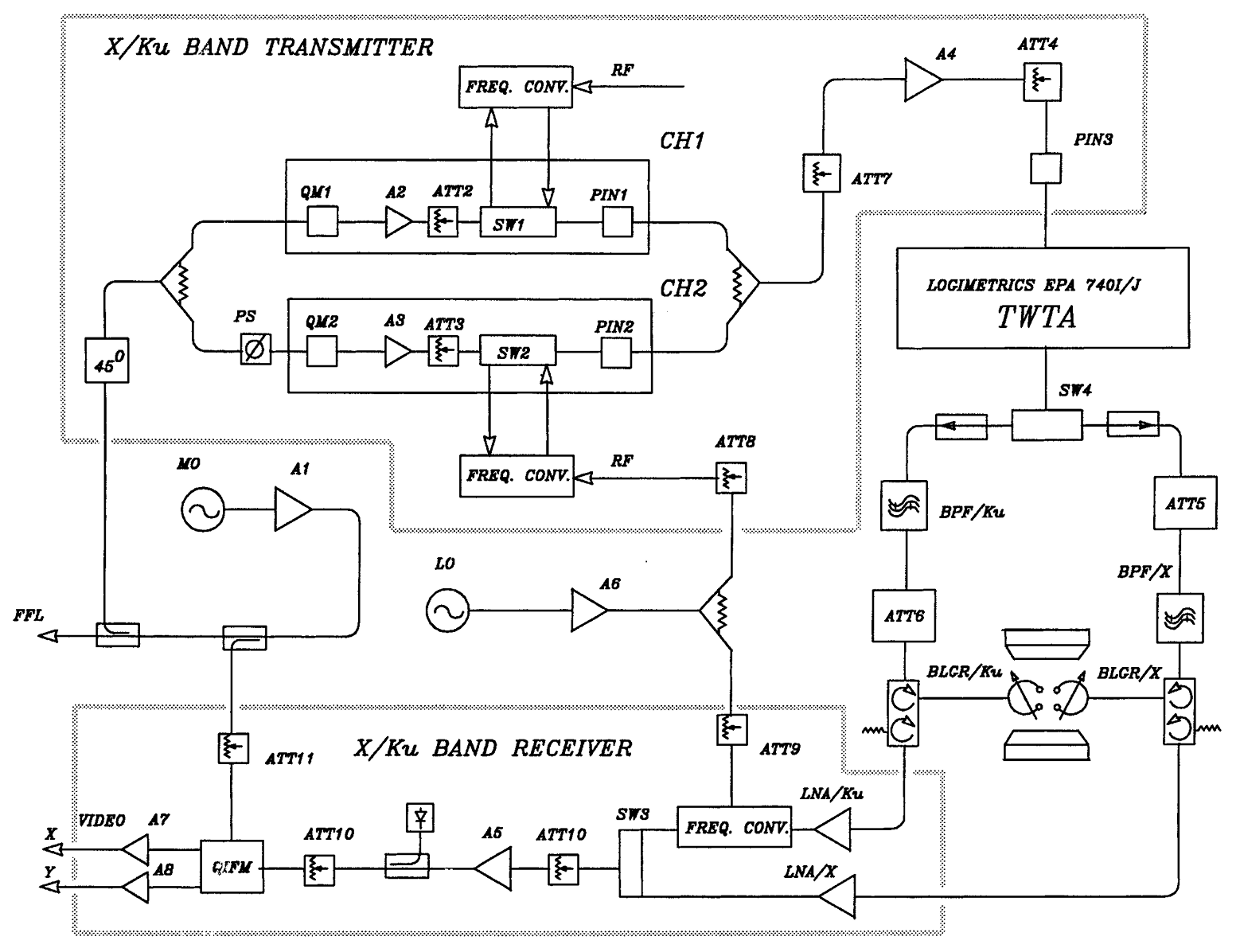
|
|
ABSTRACT: A two-dimensional Fourier Transform ESR (2D FT ESR) spectrometer operating at 9.25 and 17.35 GHz is described. The Ku-band bridge uses an efficient heterodyne technique wherein 9.25 GHz is the intermediate frequency. At Ku-band the sensitivity is increased by almost an order of magnitude. One may routinely collect a full 2D ELDOR spectrum in less than 20 min for a sample containing 0.5–5 nmol of nitroxide spin-probe in the slow-motional regime. Broad spectral coverage at Ku-band is obtained by use of a bridged loop-gap resonator (BLGR) and of a dielectric ring resonator (DR). It is shown that an even more uniform spectral excitation is obtained by using shorter microwave pulses of about 3 ns duration. The dead-time at Ku-band is just 30–40 ns, yielding an improved SNR in 2D ELDOR spectra of nitroxide spin-probes with T2 as short as 20–30 ns. A comparison of 2D ELDOR spectra obtained at 9.25 and 17.35 GHz for spin-labeled phospholipid probes (16PC) in 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol (DMPG) membrane vesicles showed that both spectra could be satisfactorily simulated using the same set of model parameters even though they are markedly different in appearance. The improved sensitivity and shorter dead-time at Ku-band made it possible to obtain orientation-dependent 2D ELDOR spectra of the Cholestane (CSL) spin-probe in macroscopically aligned lipid bilayers of egg yolk PC using samples containing only 1 mg of lipid and just 5 nmol of spin-probe.
|
|
|
Theory of Double Quantum Two-Dimensional Electron Spin Resonance with Application to Distance Measurements
S. Saxena and J.H. Freed.
J. Chem. Phys. 107, 1317-1340 (1997).
<doi: 10.1063/1.474490>
Publication #253
|
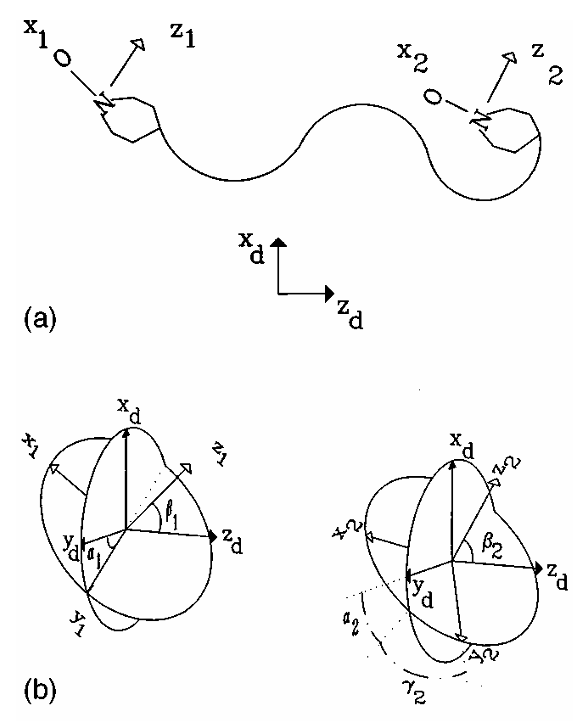
|
|
ABSTRACT: A formulation is presented for calculating double quantum two dimensional electron spin resonance (DQ-2D ESR) spectra in the rigid limit that correspond to recent experimental DQ-2D ESR spectra obtained from a nitroxide biradical. The theory includes the dipolar interaction between the nitroxide moieties as well as the fully asymmetric g and hyperfine tensors and the angular geometry of the biradical. The effects of arbitrary pulses (strong but not truly nonselective pulses) are included by adapting the recently introduced split Hamiltonian theory for numerical simulations. It is shown how arbitrary pulses in magnetic resonance create "forbidden" coherence pathways, and their role in DQ-2D ESR is delineated. The high sensitivity of these DQ-2D ESR signals to the strength of the dipolar interaction is demonstrated and rationalized in terms of the orientational selectivity of the "forbidden" pathways. It is further shown that this selectivity also provides constraints on the structural geometry (i.e., the orientations of the nitroxide moieties) of the biradicals. The theory is applied to the recent double quantum modulation (DQM) experiment on an end-labeled poly-proline peptide biradical. A distance of 18.5 Å between the ends is found for this biradical. A new two pulse double quantum experiment is proposed (by analogy to recent NMR experiments), and its feasibility for the ESR case is theoretically explored.
|
|
|
Aqueous Sample Holders for High-Frequency Electron Spin Resonance
J.P. Barnes and J.H. Freed.
Rev. Sci. Instrum. 68, 2838-2846 (1997).
<doi: 10.1063/1.1148205>
Publication #252
|
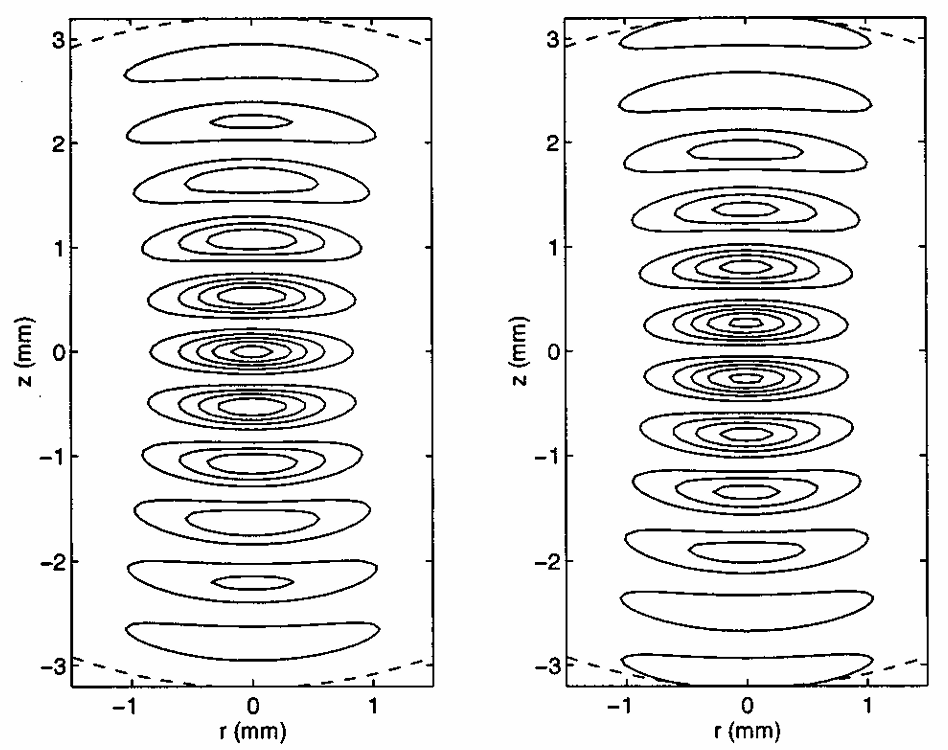
|
|
ABSTRACT: A quasioptical approach is utilized to design a resonator and sample holder for aqueous samples for high-frequency (250 GHz) electron spin resonance (ESR) spectroscopy. A disk shaped sample geometry was chosen to match the field contours of the fundamental mode in a Fabry–Perot resonator. A transmission-line analysis is used to determine the optimum sample geometry while taking into account diffractive effects of the sample and the sample holder on the far infrared (FIR) radiation field. FIR-ESR spectra of several aqueous solutions of nitroxide spin labels, including membrane vesicles and a labeled protein, are shown to demonstrate the success of the technique.
|
|
|
A 250 GHz ESR Study of o-Terphenyl: Dynamic Cage Effects Above Tc
Keith A. Earle, Jozef K. Moscicki, Antonino Polimeno, and Jack H. Freed
J. Chem. Phys. 106, 9996-10015 (1997).
<doi: 10.1063/1.474114>
Publication #251
|
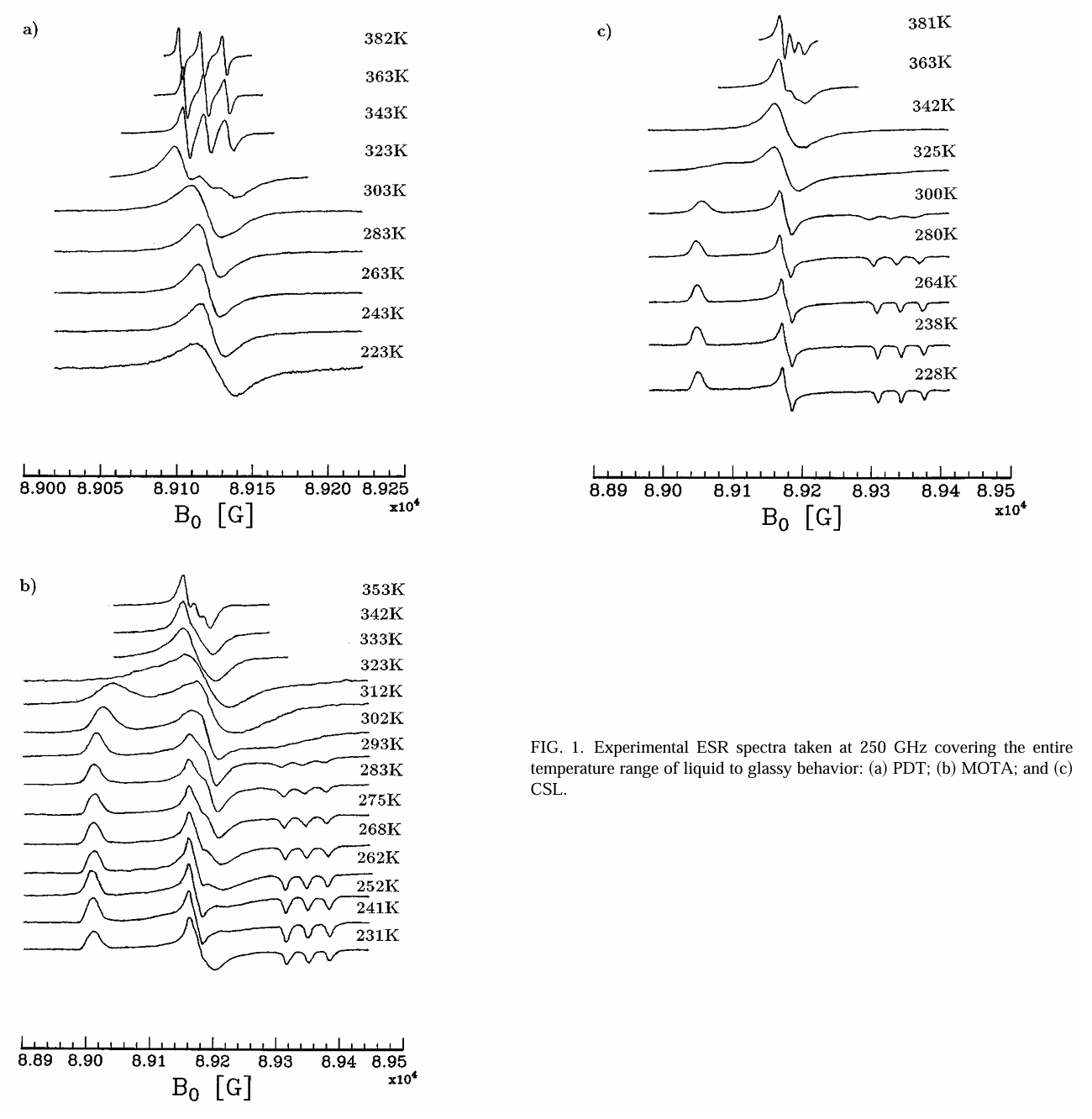
|
|
ABSTRACT: Three nitroxide spin probes of different sizes and geometrical shape were used in a 250 GHz ESR study of the probe rotational dynamics in the fragile glass former ortho-terphenyl (OTP) over a wide temperature range from 380 to 180 K. Comparative studies at 9.5 GHz have also been performed. Perdeuterated 2,2′,6,6′-tetramethyl-4-methyl aminopiperidinyl-N-oxide (MOTA), and 3,3′-dimethyloxazolidinyl-N-oxy-2′,3-5α-cholestane (CSL) are, respectively, comparable in size to and larger than the OTP host molecule, whereas Perdeuterated 2,2′,6,6′-tetramethyl-4-piperidine-N-oxide (PDT) is substantially smaller. The sensitivity of 250 GHz ESR to the details of the rotational tumbling for T≳Tc (where Tc is the crossover temperature) was exploited to show that the relaxation is fit by a model that is 2:11 PM 7/16/2025characteristic of a homogeneous liquid. A nonlinear least-squares analysis shows that below the melting point, Tm CSL, and MOTA dynamics are well-described by a model of dynamic cage relaxation proposed by Polimeno and Freed wherein the probe relaxation is significantly influenced by a fluctuating potential well created by the neighboring OTP molecules. A model of simple Brownian reorientation does not fit the experimental spectra of CSL or MOTA as well as the dynamic cage model below Tm. Spectra of PDT do not show any significant non-Brownian dynamics for this probe. It was found that the characteristic rates of the cage model, viz., the reorientation of the probe and the cage relaxation, were describable by activated processes; however, the "average" rotational diffusion rates (defined in the usual manner as the time integral of the correlation function) derived from the dynamic cage parameters follow the Stokes–Einstein–Debye (SED) relation rather well, in agreement with previous studies by other physical techniques. It is then shown that the usual stretched exponential fit to the motional correlation function, interpreted in terms of an inhomogeneous distribution of simple reorientational rates, is clearly inconsistent with the observed ESR spectrum. The absence of a significant cage potential above Tm is discussed in terms of a model of frustration limited domain sizes proposed by Kivelson and co-workers. Evidence for the existence of substantial voids in OTP below Tm especially from the spectra of the small PDT probe, is discussed in terms of the structure and packing of the OTP solvent.
|
|
|
Lipid-Gramicidin Interactions Using Two-Dimensional Fourier-Transform Electron Spin Resonance
B.R. Patyal, R.H. Crepeau, and J.H. Freed.
Biophys. J. 73, 2201-2220 (1997).
<doi: 10.1016/S0006-3495(97)78252-3>
PMID:
9336217
PMCID:
PMC1181122
Publication #250
|
|
|
ABSTRACT: The application of two-dimensional Fourier-transform electron-spin-resonance (2D-FT-ESR) to the study of lipid/gramicidin A (GA) interactions is reported. It is shown that 2D-FT-ESR spectra provide substantially enhanced spectral resolution to changes in the dynamics and ordering of the bulk lipids (as compared with cw-ESR spectra), that result from addition of GA to membrane vesicles of dipalmitoylphosphatidylcholine (DPPC) in excess water containing 16-PC as the lipid spin label. The agreement between the theory of Lee, Budil, and Freed and experimental results is very good in the liquid crystalline phase. Both the rotational and translational diffusion rates of the bulk lipid are substantially decreased by addition of GA, whereas the ordering is only slightly increased, for a 1:5 ratio of GA to lipid. The slowing effect on the diffusive rates of adding GA in the gel phase is less pronounced. It is suggested that the spectral fits in this phase would be improved with a more detailed dynamic model. No significant evidence is found in the 2D-FT-ESR spectra for a second immobilized component upon addition of GA, which is in contrast to cw-ESR. It is shown from simulations of the observed 2D-FT-ESR spectra that the additional component seen in cw-ESR spectra, and usually attributed to "immobilized" lipid, is inconsistent with its being characterized by increased ordering, according to a model proposed by Ge and Freed, but it would be consistent with the more conventional model of a significantly reduced diffusional rate. This is because the 2D-FT-ESR spectra exhibit a selectivity, favoring components with longer homogeneous relaxation times, T2. The homogeneous linewidths of the 2D-FT-ESR autopeaks appear to broaden as a function of mixing time. This apparent broadening is very likely due to the process of cooperative order director fluctuations (ODF) of the lipids in the vesicle. This real-time observation of ODF is distinct from, but appears in reasonable agreement with, NMR results. It is found that addition of GA to give the 1:5 ratio has only a small effect on the ODF, but there is a significant temperature dependence.
|
|
|
Chain Dynamics and the Simulation of Electron Spin Resonance Spectra from Oriented Phospholipid Membranes
R. Cassol, M. Ge, A. Ferrarini, and J.H. Freed.
J. Phys. Chem. B 101, 8782-8789, (1997).
<doi: 10.1021/jp970071t>
Publication #249
|
|
|
ABSTRACT: A model previously developed for describing the dynamics of flexible alkyl chains that is based on Flory's rotational isomeric state approximation is adapted and applied to the analysis of electron spin resonance (ESR) spectra obtained from a phospholipid spin label in a macroscopically aligned phospholipid membrane. In this model, rotation around each C−C bond of the labeled alkyl chain is characterized by three inequivalent minima, with one end of the chain fixed to mimic the phospholipid headgroup, and with the dynamic effects of the nitroxide label explicitly included. This model is integrated with that for the overall rotation of the phospholipid in the mean orientational potential of the aligned membrane, and it is incorporated into the stochastic Liouville equation which describes the ESR line shape in the presence of these dynamic processes. The analysis is simplified by introducing the fact that the relatively rapid internal modes of motion can be treated by motional narrowing theory and a time scale separation can be made with respect to the much slower overall motions of the phospholipid. A series of ESR spectra from the spin label 16-PC in the lipid dimyristoylphosphatidylcholine were obtained over a range of temperatures (35−65 °C) in the Lα phase for various orientations of the normal to the bilayer plane relative to the magnetic field. Very good agreement with experiment is obtained from this model by using least squares fitting procedures for the overall motional dynamics. One finds an order parameter of ❬D200❭ that is constant throughout the phase and the perpendicular component for rotational diffusion, R⊥, that ranges from about 1−3 × 107 s-1 (which corresponds to the ESR slow motional regime). Fits to the ESR spectra were also obtained from a simple but standard model wherein a single overall rotational diffusion tensor is used to describe the combined effects of internal and overall dynamics. These fits were almost as good, but they lead to a much larger R⊥ ≈ 3−6 × 108 s-1 and a smaller ❬D200❭ = 0.1, since these parameters now include the composite effects of both types of processes. New ESR experiments are proposed to provide more critical tests of these models.
|
|
|
An EPR Study of Some Highly Distorted Tetrahedral Manganese(II) Complexes at High Magnetic Fields
R.M. Wood, D.M. Stucker, L.M. Jones, W.B. Lynch, S.K. Misra, and J.H. Freed.
Inorg. Chem. 38, 5384-5388 (1999).
<doi: 10.1021/ic990377+>
Publication #248
|
|
|
ABSTRACT: EPR spectra at 94.5 GHz (3 mm) and 249.9 GHz (1 mm) have been measured for the dichloro-, dibromo-, and diiodobis(triphenylphosphine oxide)manganese(II) complexes. These compounds have a distorted tetrahedral geometry, and the highest possible symmetry obtainable is C2v. Because the spectra are taken in the high-field limit, they are simple and much easier to interpret than at lower magnetic fields. Accurate values of the zero-field splitting parameters, D and η (E∕D), are determined by least-squares fitting and homotopy using matrix diagonalization. The compounds have g-values near 2 and large values of D and η. Across the series, D increases from 0.165 to 0.906 cm-1, while η decreases slightly from 0.273 to 0.246.
|
|
|
Absorption Lineshapes in Two-Dimensional Electron Spin Resonance and the Effects of Slow Motions in Complex Fluids
S. Saxena and J.H. Freed.
J. Magn. Reson. 124, 439-454 (1997).
<doi: 10.1006/jmre.1996.1078>
PMID:
9169224
Publication #247
|
|
|
ABSTRACT: A methodology for obtaining pure absorption two-dimensional electron spin resonance spectra is presented for the case of large inhomogeneous broadening and/or slow motions. For slow motions, the spectra consist of "complex Lorentzians" superimposed with complex weighting factors, presenting a challenge to obtaining absorption spectra. It is shown how absorption-type spectra can be recovered for the two-pulse COSY and SECSY experiments in such cases. For three-pulse 2D ELDOR experiments, absorption lineshapes can be obtained for the autopeaks, whereas the cross peaks would be of mixed-mode character, in general. However, for practical cases the dispersive components in the cross peaks will be relatively small. Theoretical and experimental absorption spectra are provided to illustrate the method and to show the improved resolution obtained from absorption lineshapes. In particular, the variation in linewidths across a SECSY spectrum, which is a key component in elucidating motional dynamics, is clearly rendered in the pure absorption mode. A convenient method for introducing the necessary phase corrections for the slow-motional spectra is also provided.
|
|
|
A Theoretical Approach to the Analysis of Arbitrary Pulses in Magnetic Resonance
K.M. Salikhov, D.J. Schneider, S. Saxena, and J.H. Freed.
Chem. Phys. Lett. 262, 17-26 (1996).
<doi: 10.1016/0009-2614(96)01044-5>
Publication #246
|
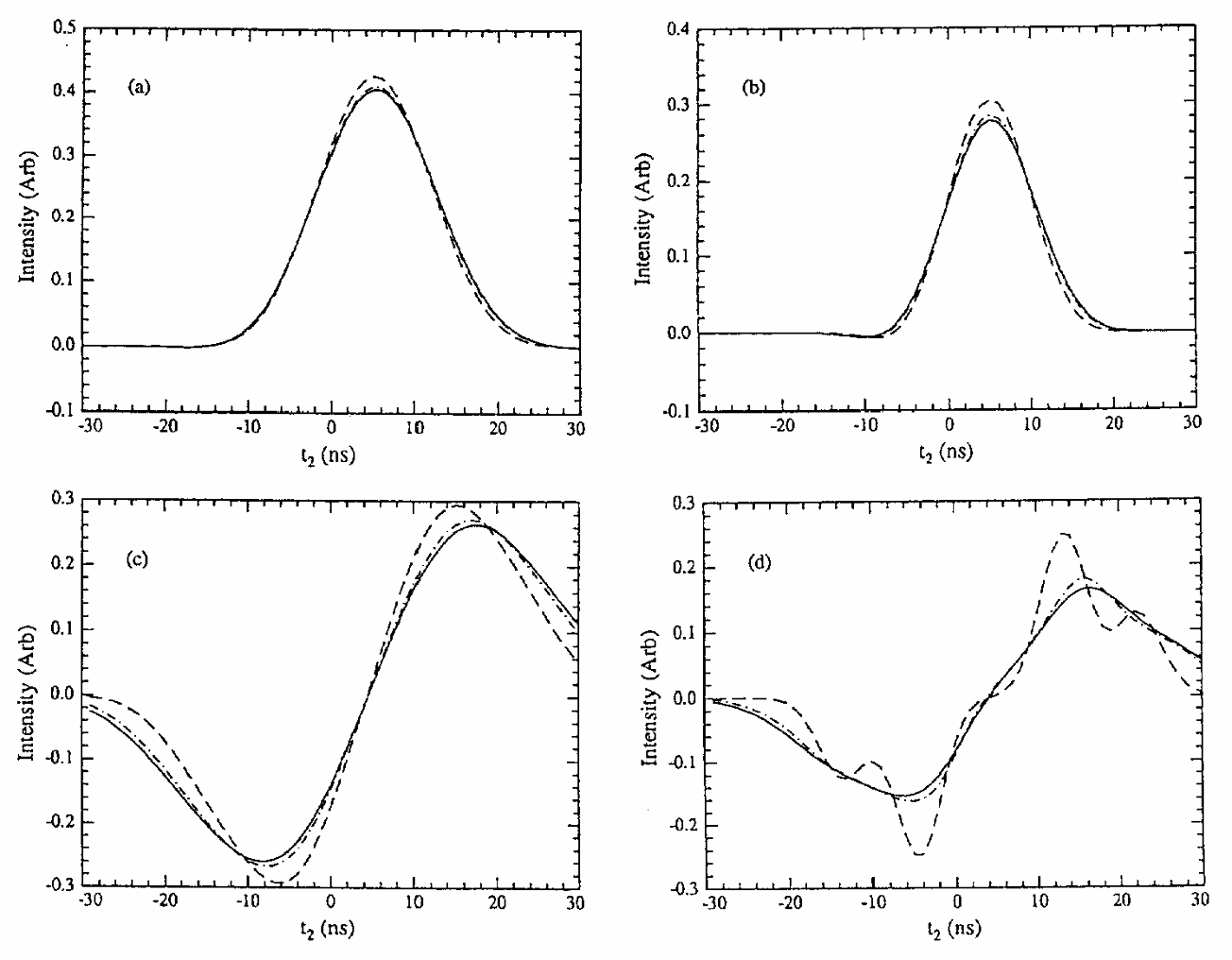
|
|
ABSTRACT: We introduce split Hamiltonian theory (SHT) to analyze arbitrary pulses in magnetic resonance, i.e. pulses substantial in magnitude but not non-selective. The range of validity of the lower order approximations is discussed, and the method is illustrated by applying it to the consideration of pulse adjustable spectroscopies in time domain ESR that are utilized to study nuclear modulation. A virtue of SHT is that, whereas the approximate analytic solutions can provide useful insights, it can also be iterated numerically to achieve quantitatively accurate solutions
|
|
|
Studies of Spin Relaxation and Molecular Dynamics in Liquid Crystals by Two-Dimensional Fourier Transform Electron Spin Resonance. II. Perdeuterated-Tempone in Butoxy Benzylidene Octylaniline and Dynamic Cage Effects
V.S.S. Sastry, A. Polimeno, R.H. Crepeau, and J.H. Freed.
J. Chem. Phys. 105, 5773-5791 (1996).
<doi: 10.1063/1.472421>
Publication #245
|
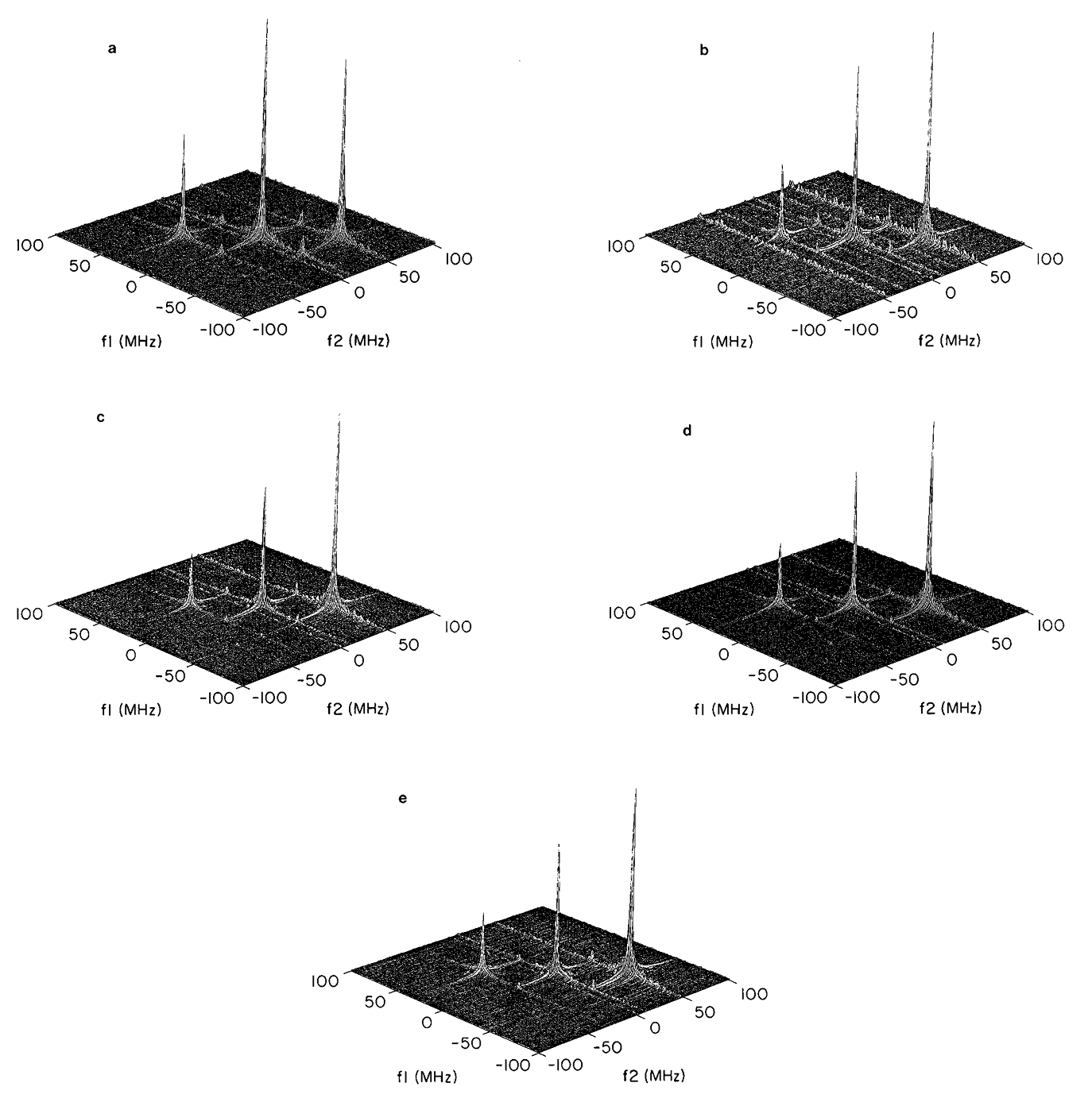
|
|
ABSTRACT: Two-dimensional Fourier transform (2D-FT)-electron spin resonance (ESR) studies on the small globular spin probe perdeuterated tempone (PDT) in the liquid crystal solvent 4O,8 (butoxy benzylidene octylaniline) are reported. These experiments, over the temperature range of 95 °C to 24 °C, cover the isotropic (I), nematic (N), smectic A (SA), smectic B (SB), and crystal (C) phases. The 2D-ELDOR (two-dimensional electron–electron double resonance) spectra confirm the anomalously rapid reorientation of PDT, especially in the lower temperature phases. The model of a slowly relaxing local structure (SRLS) leads to generally very good non-linear least squares (NLLS) global fits to the sets of 2D-ELDOR spectra obtained at each temperature. These fits are significantly better than those achieved by the standard model of Brownian reorientation in a macroscopic orienting potential. The SRLS model is able to account for anomalies first observed in an earlier 2D-ELDOR study on PDT in a different liquid crystal in its smectic phases. Although it is instructional to extract the various spectral densities from the COSY (correlation spectroscopy) and 2D-ELDOR spectra, the use of NLLS global fitting to a full set of 2D-ELDOR spectra is shown to be more reliable and convenient for obtaining optimum model parameters, especially in view of possible (incipient) slow motional effects from the SRLS or dynamic cage. The cage potential is found to remain fairly constant at about kBT over the various phases (with only a small drop in the SB phase), but its asymmetry increases with decreasing temperature T. This value is significantly larger than the weak macroscopic orienting potential which increases from 0.1 to 0.3kBT with decreasing T. The cage relaxation rate, given by Rc is about 3×107 s−1 in the I phase, but increases to about 108 s−1 in the SA, SB, and C phases. The rotational diffusion tensor for PDT shows only a small T-independent asymmetry, and its mean rotational diffusion coefficient is of order 1010 s−1, with however, a small increase in the SB phase. These results are consistent with a model previously proposed for PDT in benzylidene liquid crystal solvents, that as T is reduced the PDT molecules are partially expelled from the hard core (dipolar) region of the liquid crystalline molecules toward the more flexible aliphatic chain region as a result of increased core packing from smectic layer formation, and it thus experiences a more fluid (for a given temperature) local cage structure.
|
|
|
Studies of Spin Relaxation and Molecular Dynamics in Liquid Crystals by Two-Dimensional Fourier Transform Electron Spin Resonance. I. Cholestane in Butoxy Benzylidene-Octylaniline and Dynamic Cage Effects
V.S.S. Sastry, A. Polimeno, R.H. Crepeau, and J.H. Freed.
J. Chem. Phys. 105, 5753-5772 (1996).
<doi: 10.1063/1.472420>
Publication #244
|
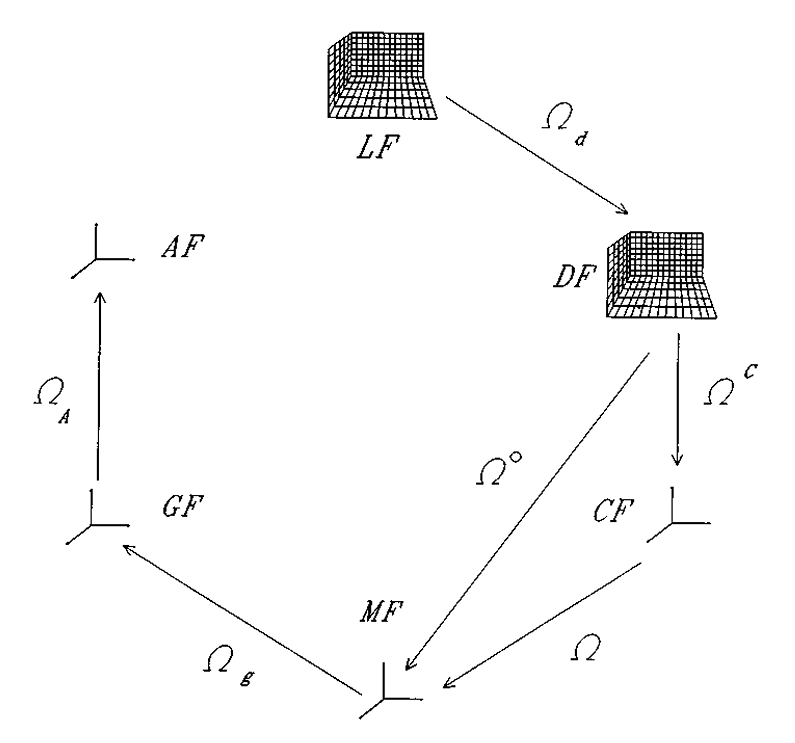
|
|
ABSTRACT: Two-dimensional Fourier transform (2D-FT) electron spin resonance (ESR) studies on the rigid rodlike cholestane (CSL) spin-label in the liquid crystal solvent 4O,8 (butoxy benzylidene octylaniline) are reported. These experiments were performed over a wide temperature range: 96 °C to 25 °C covering the isotropic (I), nematic (N), smectic A (SA), smectic B (SB), and crystal (C) phases. It is shown that 2D-FT-ESR, especially in the form of 2D-ELDOR (two-dimensional electron–electron double resonance) provides greatly enhanced sensitivity to rotational dynamics than previous cw-ESR studies on this and related systems. This sensitivity is enhanced by obtaining a series of 2D-ELDOR spectra as a function of mixing time, Tm, yielding essentially a three-dimensional experiment. Advantage is taken of this sensitivity to study the applicability of the model of a slowly relaxing local structure (SRLS). In this model, a dynamic cage of solvent molecules, which relaxes on a slower time scale than the CSL solute, provides a local orienting potential in addition to that of the macroscopic aligning potential in the liquid crystalline phase. The theory of Polimeno and Freed for SRLS in the ESR slow motional regime is extended by utilizing the theory of Lee et al. to include 2D-FT-ESR experiments, and it serves as the basis for the analysis of the 2D-ELDOR experiments. It is shown that the SRLS model leads to significantly improved non-linear least squares fits to experiment over those obtained with the standard model of Brownian reorientation in a macroscopic aligning potential. This is most evident for the SA phase, and the use of the SRLS model also removes the necessity of fitting with the unreasonably large CSL rotational asymmetries in the smectic phases that are required in both the cw-ESR and 2D-ELDOR fits with the standard model. The cage potential is found to vary from about kBT in the isotropic phase to greater than 2kBT in the N and SA phases, with an abrupt drop to about 0.2kBT in the SB and C phases. Concomitant with this drop at the SA–SB transition is an almost comparable increase in the orienting potential associated with the macroscopic alignment. This is consistent with a freezing in of the smectic structure at this transition. The cage relaxation rate given by Rc, its "rotational diffusion coefficient," is of order of 107 s−1 in the I and N phases. It drops somewhat in the SA phase, but there is a greater than order of magnitude drop in Rc for the SB and C phases to about 105 s−1. This drop is also consistent with the freezing in of the smectic structure. The rotational diffusion tensor of the CSL probe is significantly larger than Rc which is consistent with the basic physical premise of the SRLS model. In particular, R⊥o and R∥o are of order 108 s−1 and 109 s−1 respectively.
|
|
|
Far-Infrared Electron-Paramagnetic-Resonance Spectrometer Utilizing a Quasioptical Reflection Bridge
K.A. Earle, D.S. Tipikin, and J.H. Freed.
Rev. Sci. Instrum. 67, 2502-2513 (1996).
<doi: 10.1063/1.1147205>
Publication #243
|
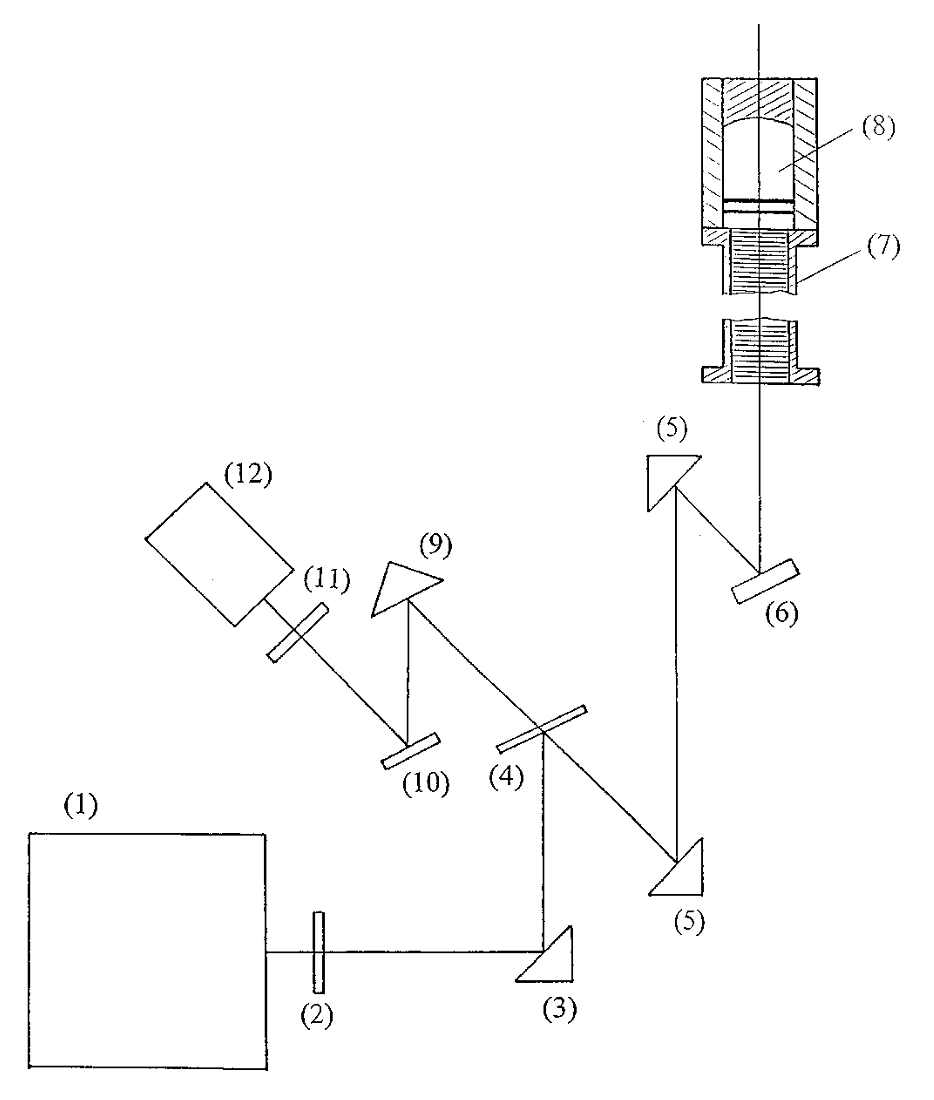
|
|
ABSTRACT: We describe a far-infrared electron-paramagnetic-resonance (EPR) spectrometer for broadband (100–300 GHz) use. The spectrometer is operated in the reflection mode and uses broadband quasioptical methods to separate the transmitted from the reflected radiation. We describe and illustrate its operation at 170 GHz (1.8 mm) and compare its performance to that of our transmission mode spectrometer operating at 250 GHz. We also discuss the advantages of the reflection bridge for performing EPR experiments over a broad range of frequencies, and we consider methods of improving the performance of the bridge. This includes a novel design for variable coupling of the reflection-mode Fabry–Pérot resonator.
|
|
|
The High-Frequency EPR Spectra of Polyaniline: Line Narrowing due to the Spin Exchange
D.S. Tipikin, K.A. Earle, J.H. Freed.
Polym. Sci. B 41, 151-154 (1999).
Publication #242
|
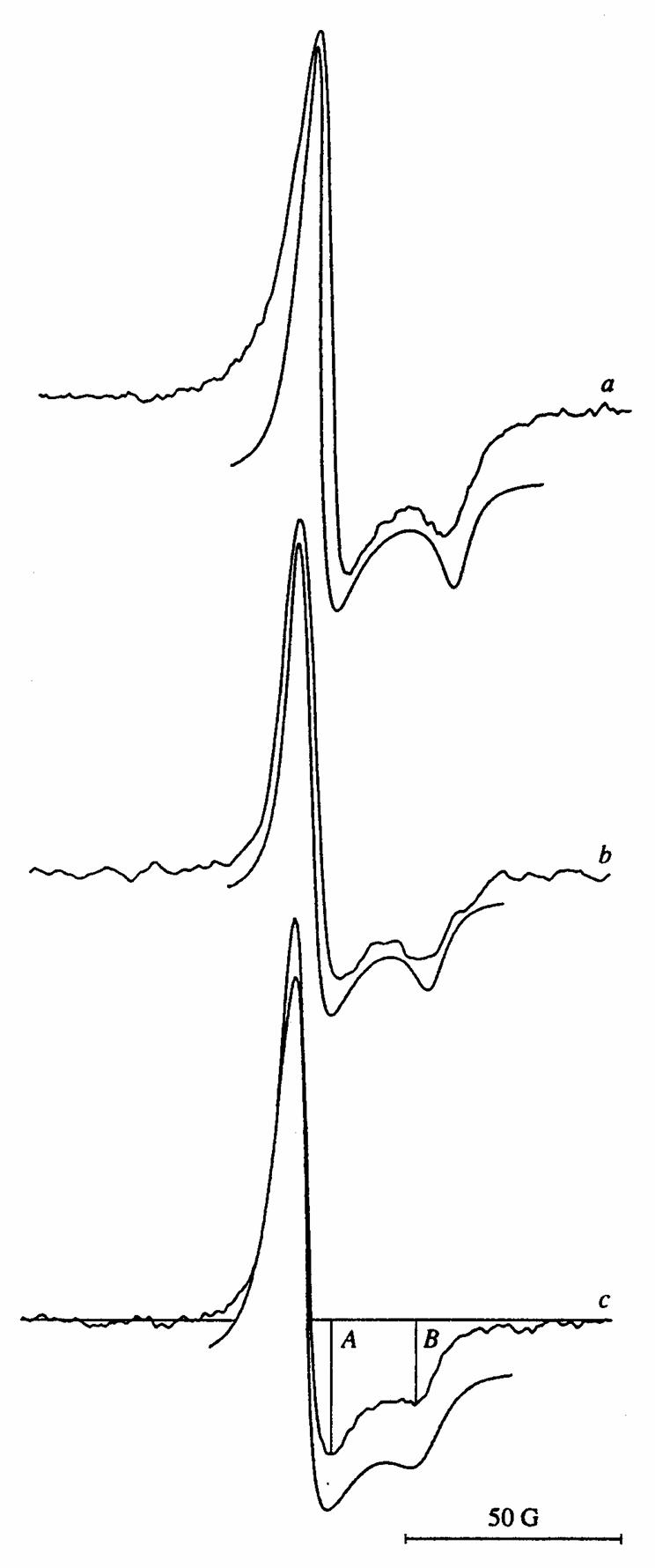
|
|
ABSTRACT: High-field EPR spectroscopy at 250 and 170 GHz was used to study polyaniline (PA) in the 100–300 K temperature range. Splitting of the EPR spectrum was observed which depended linearly on the magnetic field strength. The line shape for a PA sample was found to depend on the temperature.The spectra were simulated using the dynamic model of spin exchange.
|
|
|
Molecular Dynamics of a Liquid Crystalline Polymer Studied by Two-Dimensional Fourier Transform and CW ESR
D. Xu, R.H. Crepeau, C.K. Ober, and J.H. Freed.
J. Phys. Chem. 100, 15873-15885 (1996).
<doi: 10.1021/jp9605156>
Publication #241
|
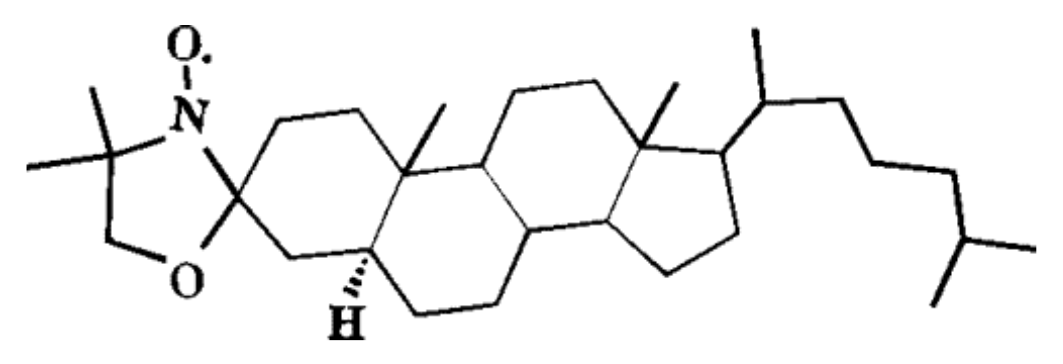
|
|
ABSTRACT: Two-dimensional Fourier transform (2D-FT) and CW-ESR experiments at X-band frequencies were performed over a broad range of temperatures covering the solid and melt states of a liquid crystalline (LC) polymer. The CW-ESR experiments were analyzed by conventional motional models. The nematic phase was macroscopically aligned in the dc magnetic field, whereas the solid state showed microscopic order but macroscopic disorder (MOMD). An end-label on the polymer showed smaller ordering and larger reorientational rates than that of the cholestane (CSL) spin probe dissolved in the same polymer, since the former can reorient by local internal chain modes. It was demonstrated that the 2D-FT-ESR experiments provide greatly enhanced resolution to the ordering and dynamics of the end-label, especially when performed as 2D-ELDOR (electron−electron double resonance) experiments as a function of mixing time. The conventional model of Brownian reorientation in an orienting potential was unsuccessful in interpreting these results. Instead the model of a slowly relaxing local structure (SRLS), which enables differentiation between the local internal modes experienced by the end-label and the collective reorganization of the polymer molecules around the end label, yielded much improved fits to the experiments in the nematic phase. These nonlinear least squares fits showed that the polymer "cage" relaxes more than 2 orders of magnitude slower than the local end-chain modes, and there is a moderate orientational potential coupling the local end-chain motion to the "cage" with axial and nonaxial local order parameters of about 0.14 and 0.29 at 100 °C. In the solid state the 2D-ELDOR spectra were fitted to the MOMD model, which is a limiting case of the SRLS model when the cage relaxation becomes very slow.
|
|
|
Rotational Diffusion and Order Parameters of a Liquid Crystalline Polymer Studied by ESR: Molecular Weight Dependence
D. Xu, D.E. Budil, C.K. Ober, and J.H. Freed.
J. Phys. Chem. 100, 15867-15872 (1996).
<doi: 10.1021/jp960514d>
Publication #240
|
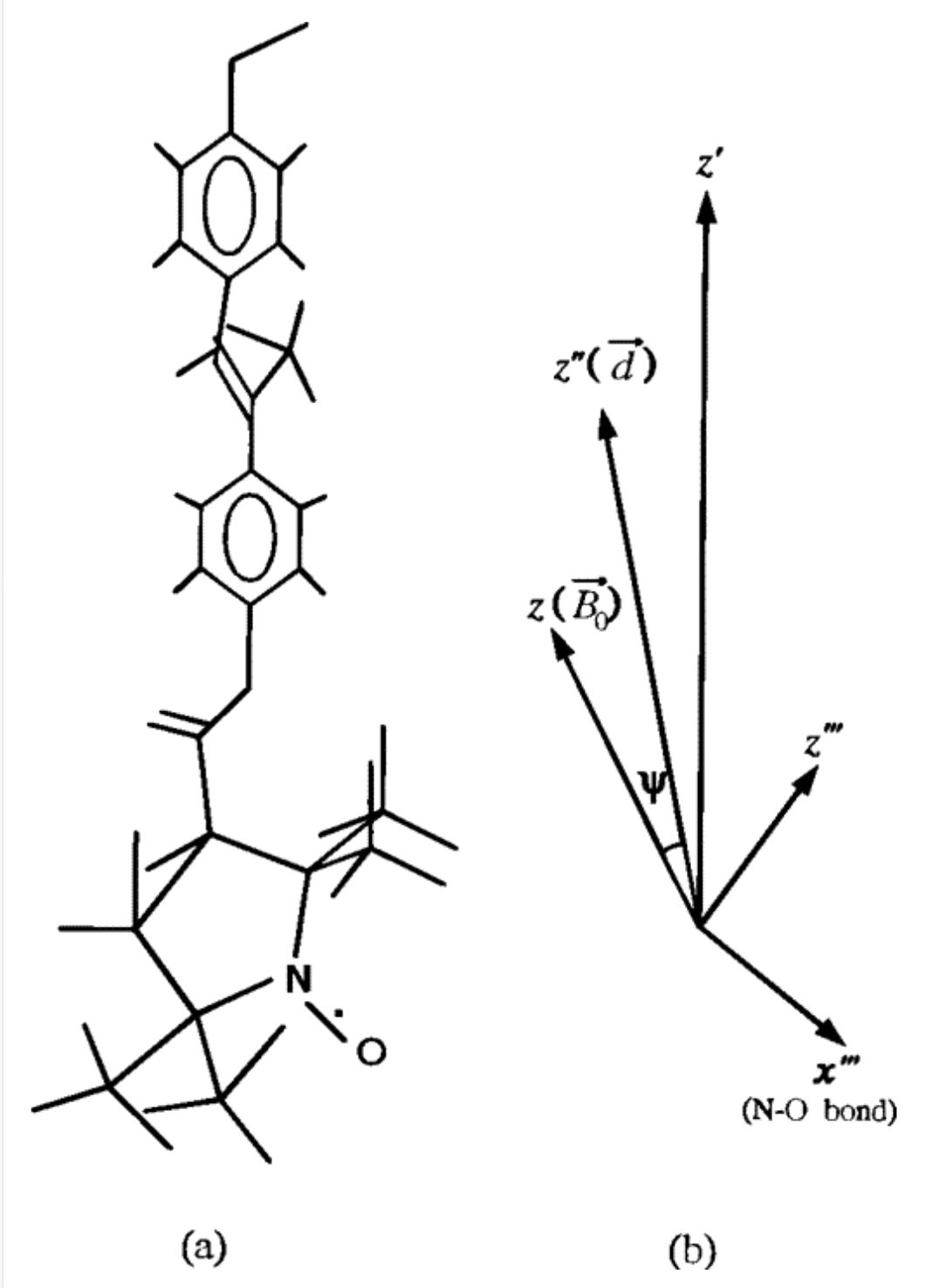
|
|
ABSTRACT: The microscopic rotational dynamics of a main chain liquid crystalline (LC) poly(ether) in its nematic phase is studied in detail by nonlinear least squares analysis of ESR spectra in the slow motional regime. This complements results reported in an accompanying paper, which focuses on macroscopic translational diffusion using the DID-ESR (dynamic imaging of diffusion by ESR) technique. Far infrared 250 GHz ESR spectroscopy is used to determine the magnetic g and A tensors of the 3-carboxy-PROXYL spin label attached to the LC polymer. ESR spectra of the labeled polymers of varying molecular weights are analyzed to yield the rotational diffusion coefficients and orientational order parameters. Different cases of the degree of macroscopic alignment are observed in these samples and accounted for in the simulations. For molecular weights lower than 11 000 (for both tracers and matrices), the rotational diffusion coefficient R̄ is found to correlate with the molecular weight of the polymer matrix and to be independent of the molecular weight of tracer, suggesting the importance of free volume for end-chain motion. Macroscopically aligned samples, corresponding to lower molecular weight LC polymers, show an inverse correlation of R̄ with order parameter, consistent with observations previously reported for nonpolymeric LCs, which were associated with free volume effects.
|
|
|
Translational Diffusion in Polydisperse Polymer Samples Studied by Dynamic Imaging of Diffusion ESR
D. Xu, E. Hall, C.K. Ober, J.K. Moscicki, and J.H. Freed.
J. Phys. Chem. 100, 15856-15866 (1996).
<doi: 10.1021/jp960513l>
Publication #239
|
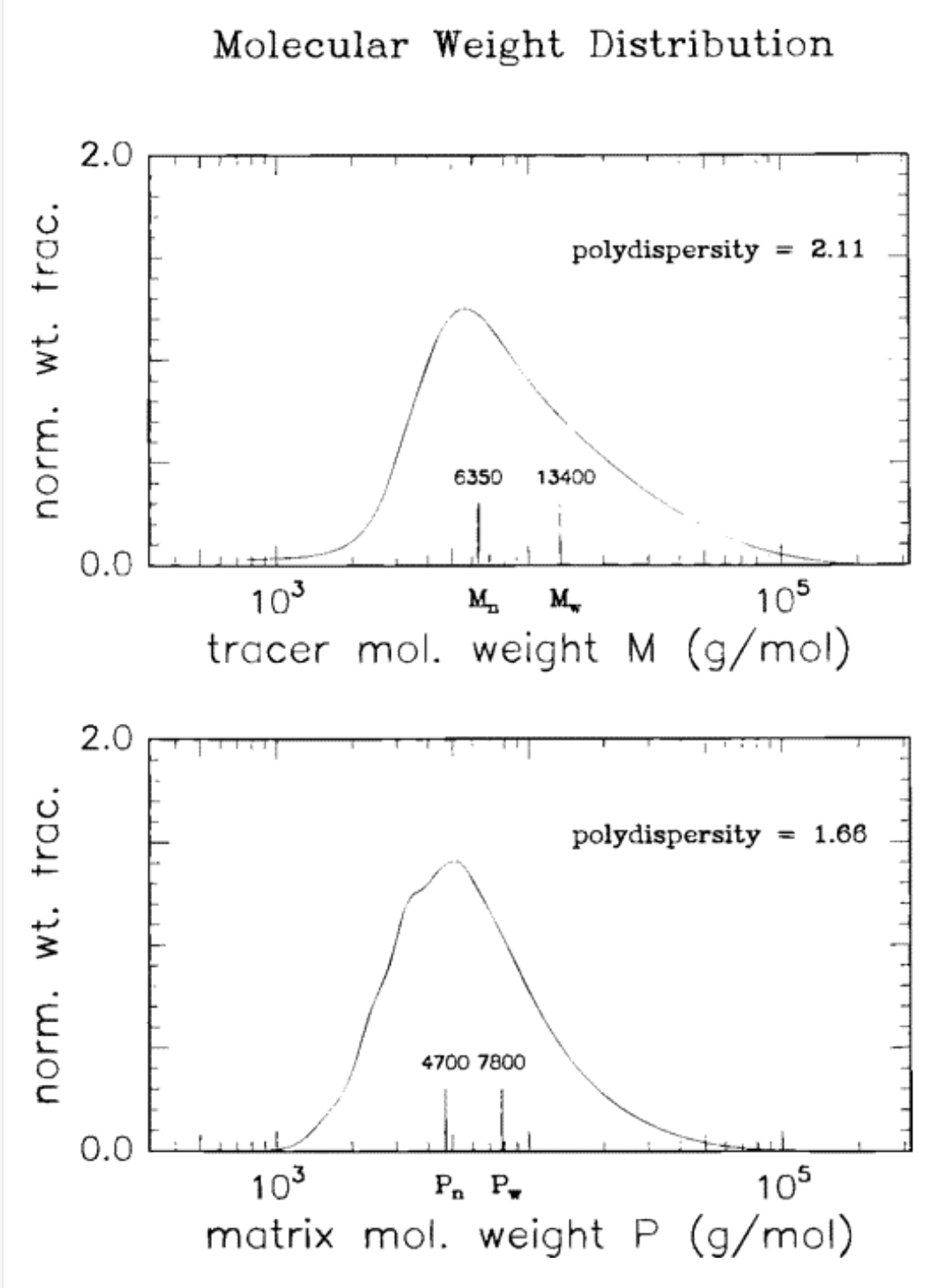
|
|
ABSTRACT: A new methodology utilizing the DID-ESR (dynamic imaging of diffusion by ESR) technique has been developed for its application to polydisperse polymer samples. Due to polydispersity the spin-labeled polymer molecules have a wide range of molecular weight, and this is also true for the matrix polymer molecules. This generally leads to a wide distribution in diffusion coefficients of the spin-labeled molecules. The work presented here includes a theoretical derivation of the new DID-ESR method in the presence of polydispersity, plus its first application for measuring translational diffusion coefficients of liquid crystalline (LC) polymer melts. This includes a detailed analysis of the reliability of the method, the proper interpretation of average diffusion coefficients, and how the molecular weight dependence of the diffusion coefficient may be obtained from even a single experiment on a sample with wide polydispersity. The results obtained by this method are compared with those from the FRES (forward recoil spectroscopy) technique on the same model system, but for higher molecular weight, and account is taken of the differences in the two methods. Future experiments to further study LC polymer diffusion mechanisms are proposed.
|
|
|
Double Quantum Two-Dimensional Fourier Transform Electron Spin Resonance: Distance Measurements
S. Saxena and J.H. Freed.
Chem. Phys. Lett. 251, 102-110 (1996).
<doi: 10.1016/0009-2614(96)00075-9>
Publication #238
|
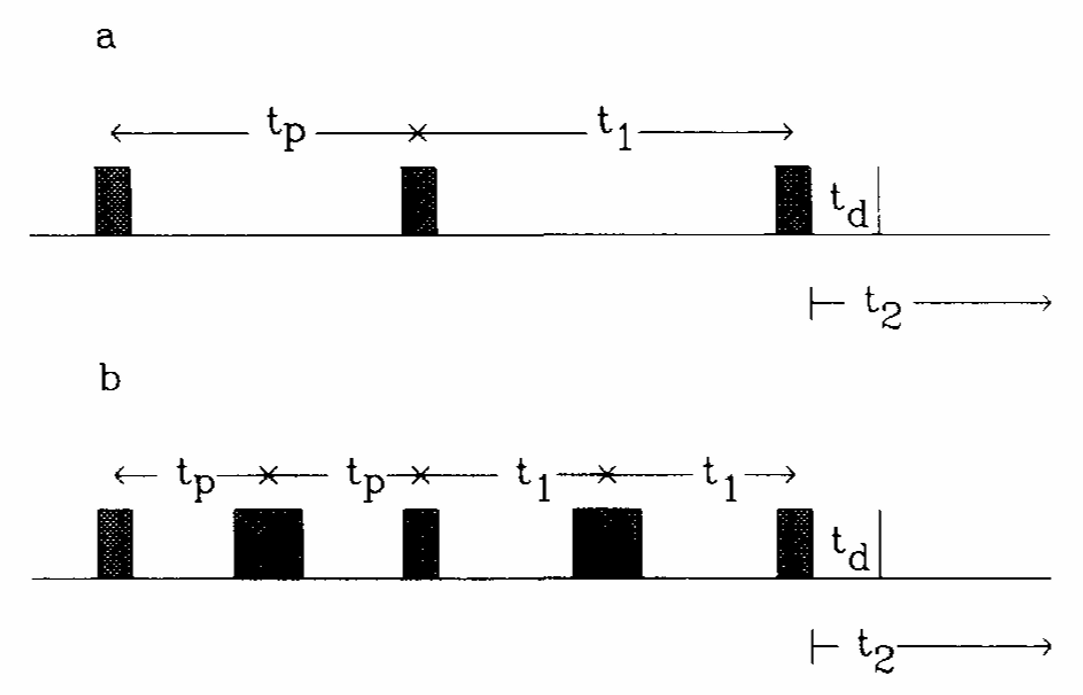
|
|
ABSTRACT: The first double quantum two-dimensional Fourier transform electron spin resonance (2D-FT ESR) experiments are reported. Extension of 2D-FT ESR to enable detection of the ΔMs = ±2 transition enhances the capability of ESR in studying structural properties (i.e. distances in bilabeled molecules). A distance of 21 Å was found for a poly-proline peptide, spin labeled at both ends, in agreement with earlier measurements using fluorescence energy transfer.
|
|
|
|
|
ESR Spectrometer at 250GHz
K.A. Earle and J.H. Freed.
SPIE 2558, 86-97 (1995).
<doi: 10.1117/12.224225>
Publication #236
|
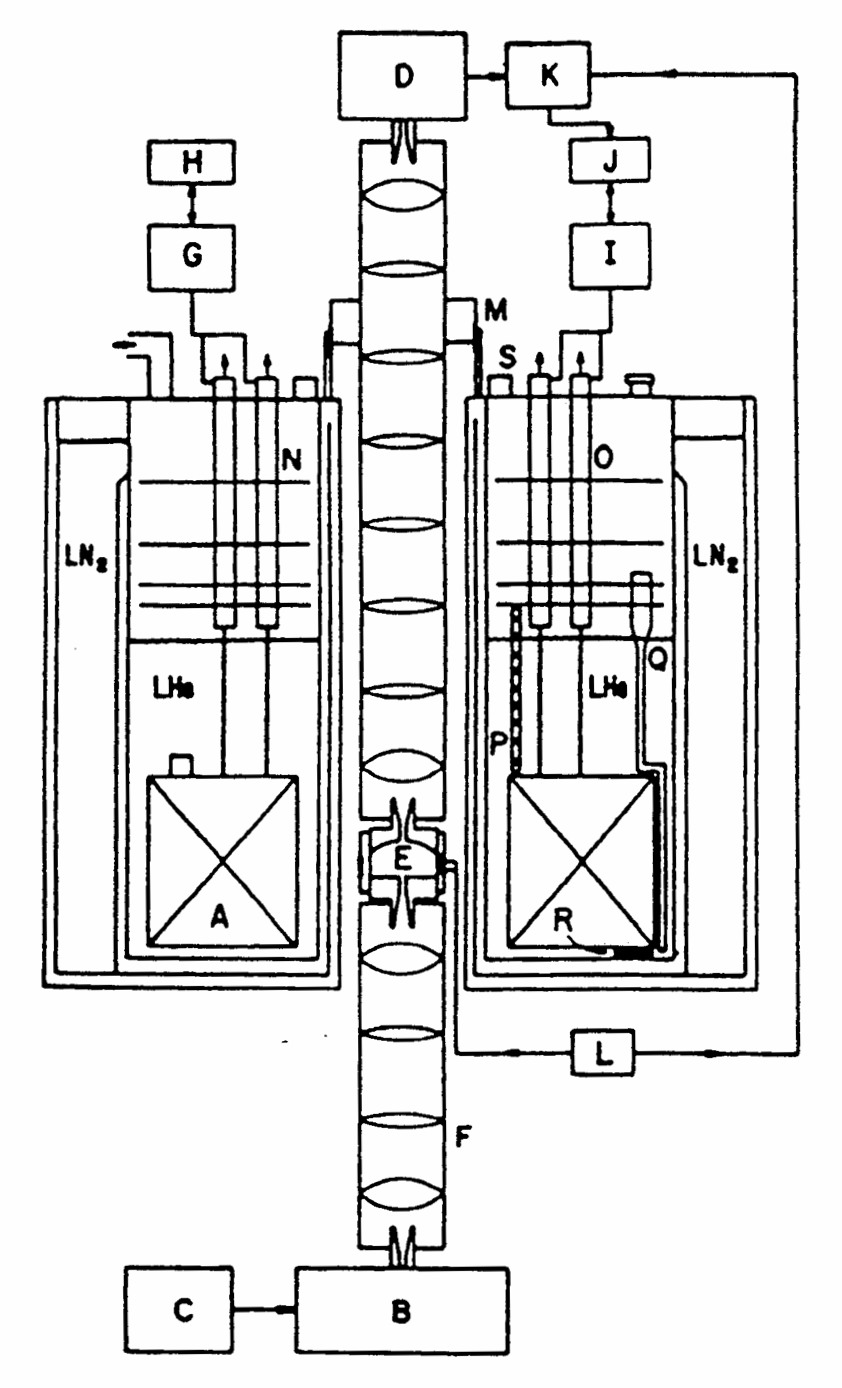
|
|
ABSTRACT: We describe the design principles of an electron spin resonance spectrometer operating at 250GHz, which uses quasioptics to propagate the far-infrared radiation instead of conventional waveguide techniques. We include examples that illustrate the sensitivity and flexibility of the spectrometer. We also include a quasi-optical analysis of the expected performance of a novel reflection mode spectrometer.
|
|
|
Site Selective Electron Paramagnetic Resonance Study of Photoexcited Chromium Doped Forsterite
A. Regev and J.H. Freed.
J. Chem. Phys. 103, 5315-5325 (1995).
<doi: 10.1063/1.470566>
Publication #235
|
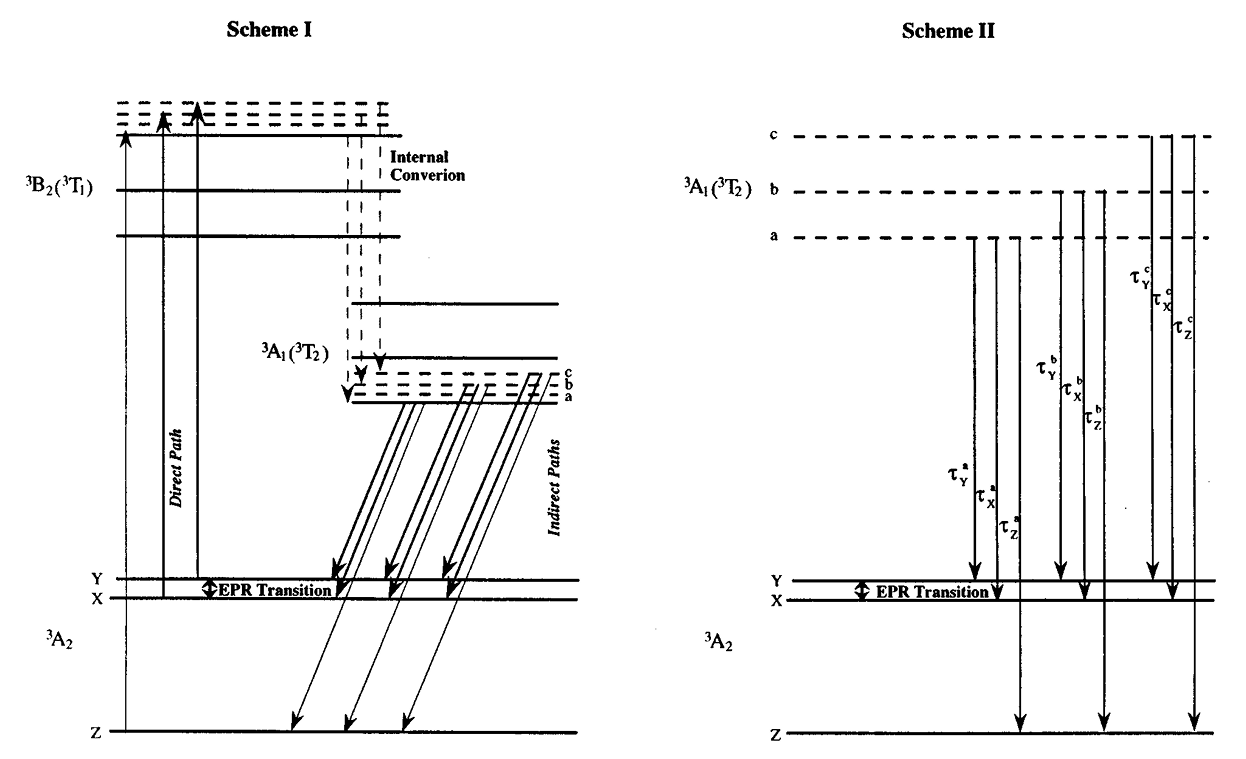
|
|
ABSTRACT: Time resolved electron paramagnetic resonance (EPR) measurements on a photoexcited chromium doped forsterite (Cr:Fo) single crystal are reported. The spectral changes with time, magnetic field, crystal orientation, microwave power and, in particular, photoexciting wavelength, provide a selective picture of the various chromium dopants and the absorption–relaxation cycle associated with each optical excitation. Both Cr+4 ions lodged at tetrahedral (Td) and octahedral (Oh) sites as well as Cr+3 ions are detected. In particular, the laser-EPR technique enabled us to monitor the spin dynamics associated with the lasing center (Cr+4∕Td) in the time regime of 200 ns–100 ms following a selective photoexcitation of the crystal between 532 and 1064 nm. The transient EPR signals associated with the lasing Cr+4∕Td ions, exhibit a noticeable dependence on even small changes (˜0.5 nm) in the exciting wavelengths that correspond to the visible 3A2→3T1 and the near infrared 3A2→3T2 transitions. The transient magnetization associated with each absorption–relaxation cycle is quantitatively analyzed in terms of site selectivity due to the narrow band (i.e., low intensity) microwave detection following a narrow band optical excitation. Given this observed selectivity, it is suggested that laser-EPR may be employed to study intersite interactions and site structure versus optical function relationships in forsterite as well as other solids doped with transition metal ions.
|
|
|
ESR Studies of Nitrogen Oxides Adsorbed on Zeolite Catalysts: Analysis of Motional Dynamics
H. Yahiro, M. Shiotani, J.H. Freed, M. Lindgren, and A. Lund.
In Catalysis by Microporous Materials, H.K. Beyer, H.G. Karge, I. Kiricsi, and J.B. Nagy, Eds.
Studies in Surface Science and Catalysis Volume 94, pp 673-680 (1995).
|
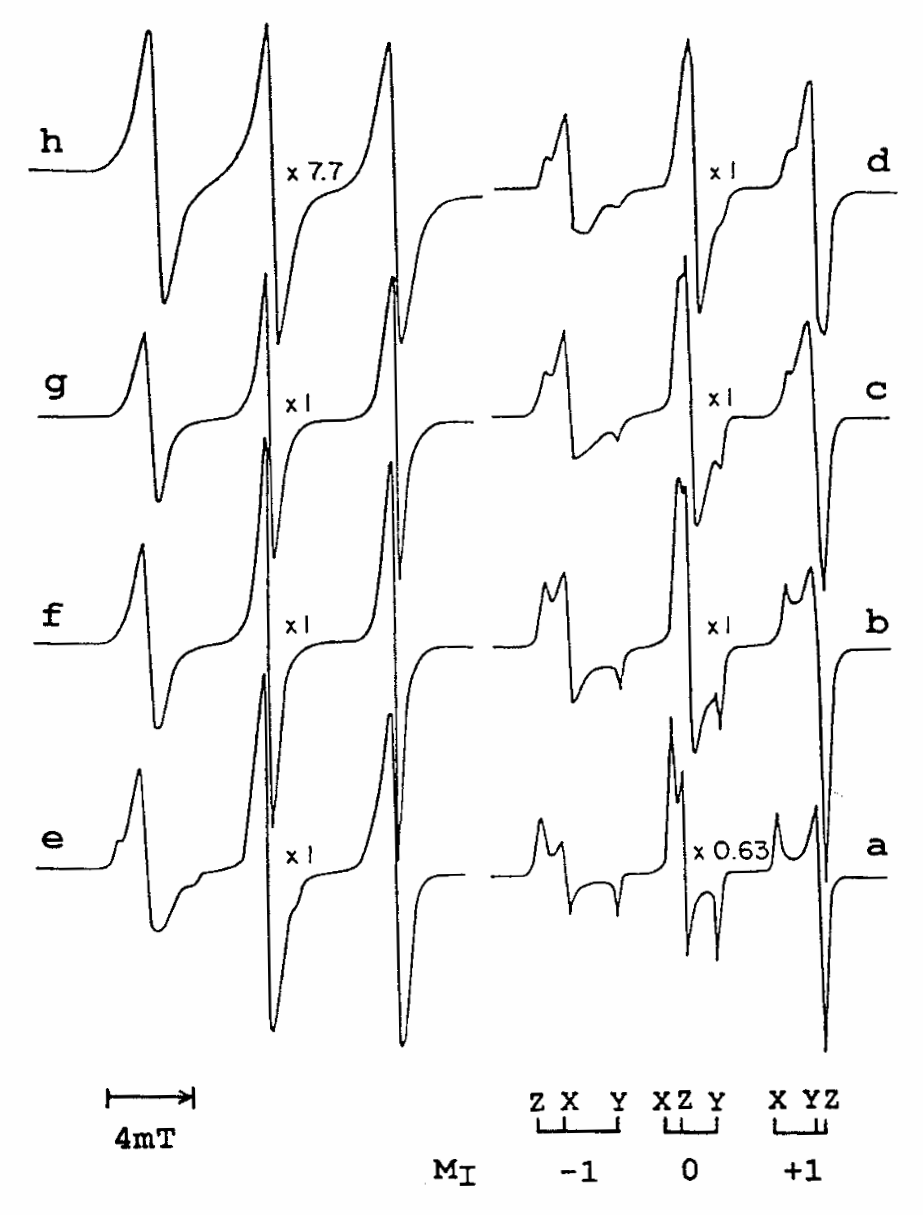
|
|
ESR Studies of Nitrogen Oxides Adsorbed on Zeolite Catalysts: Analysis of Motional Dynamics
H. Yahiro, M. Shiotani, J.H. Freed, M. Lindgren, and A. Lund.
In Catalysis by Microporous Materials, H.K. Beyer, H.G. Karge, I. Kiricsi, and J.B. Nagy, Eds.
Studies in Surface Science and Catalysis Volume 94, pp 673-680 (1995).
<doi: 10.1016/S0167-2991(06)81283-8>
Publication #234
|

|
|
ABSTRACT: ESR spectra of NO2 adsorbed on X- and Y-type zeolites were observed in the temperature range 77–346 K. Based upon spectral simulation using a Brownian diffusion model, motional dynamics of NO2 adsorbed on zeolite surface were analyzed quantitatively. In the case of X-type zeolite, it was found that the ESR spectra below 100 K is near the rigid limit. Above 230 K, the average rotational correlation time decreased from 1.7×10−9 (230 K) to 7.5×10−10sec (325 K) with increasing temperature and its degree of anisotropy was very close to one (N=1.25). On the other hand, the temperature-dependent ESR spectra of NO2 adsorbed on Y-type zeolite were observed to be somewhat different from that for X-type zeolite.
|
|
|
Slow Motional ESR in Complex Fluids: The Slowly Relaxing Local Structure Model of Solvent Cage Effects
A. Polimeno and J.H. Freed.
J. Phys. Chem. 99, 10995-11006 (1995).
<doi: 10.1021/j100027a047>
Publication #233
|
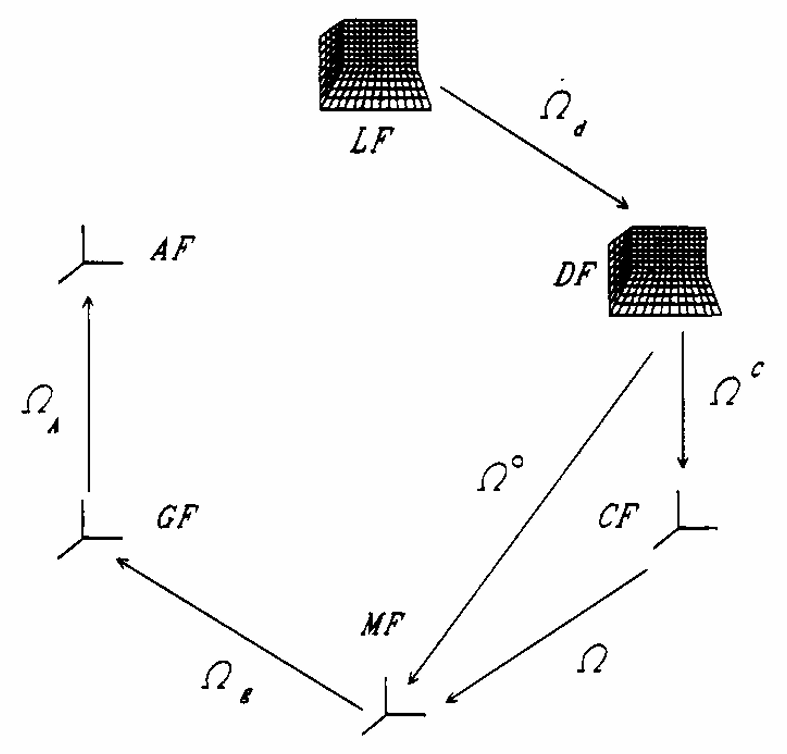
|
|
ABSTRACT: A detailed formulation is presented for the analysis of slow motional ESR in terms of the reorientation of the probe molecule within a dynamic solvent cage. This formulation is appropriate for isotropic and ordered fluids. The solvent cage is modeled in terms of a set of collective variables that represent the instantaneous solvent structure around the probe and that reorient on a slower time scale than the probe. This "slowly relaxing local structure" model is incorporated into an augmented stochastic Liouville equation that is solved by efficient computational means which enables nonlinear least squares fitting to experimental spectra. This formulation is applied to some recent slow motional ESR spectra obtained at 250 GHz. Such high-frequency ESR spectra have been shown to be particularly sensitive to the microscopic details of the molecular reorientational process. Significant improvements are found in fitting the ESR spectra for the cases studied, viz., perdeuterated 2,2,6,6-tetramethyl-4-piperidone (PDT) in toluene and 3-doxylcholestane (CSL) in o-terphenyl (OTP), a glass-forming liquid, when compared to a model of simple Brownian reorientation. In both cases the cage is found to relax at least 1 order of magnitude slower than the probe itself, and it provides a potential for probe reorientation on the order of 2-7 kBT. The cage potential for the PDT case is characterized by minima at more than one orientational angle, allowing for jump-type reorientations between such minima superimposed on substantial local motions suggestive of earlier simulations based on a simple jump model. For CSL in OTP, weak negative ordering is found, consistent with an oblate-shaped local structure provided by the OTP solvent molecules. These examples illustrate the potential of utilizing high-frequency slow motional ESR to discern details of solvent interactions associated with molecular reorientations in fluids.
|
|
|
Nonlinear-Least-Squares Analysis of Slow-Motion EPR Spectra in One and Two Dimensions Using a Modified Levenberg-Marquardt Algorithm
D.E. Budil, S. Lee, S. Saxena, and J.H. Freed.
J. Magn. Reson. A 120, 155-189 (1996).
<doi: 10.1006/jmra.1996.0113>
Publication #232
|
|
|
ABSTRACT: The application of the "model trust region" modification of the Levenberg–Marquardt minimization algorithm to the analysis of one-dimensional CW EPR and multidimensional Fourier-transform (FT) EPR spectra especially in the slow-motion regime is described. The dynamic parameters describing the slow motion are obtained from least-squares fitting of model calculations based on the stochastic Liouville equation (SLE) to experimental spectra. The trust-region approach is inherently more efficient than the standard Levenberg–Marquardt algorithm, and the efficiency of the procedure may be further increased by a separation-of-variables method in which a subset of fitting parameters is independently minimized at each iteration, thus reducing the number of parameters to be fitted by nonlinear least squares. A particularly useful application of this method occurs in the fitting of multicomponent spectra, for which it is possible to obtain the relative population of each component by the separation-of-variables method. These advantages, combined with recent improvements in the computational methods used to solve the SLE, have led to an order-of-magnitude reduction in computing time, and have made it possible to carry out interactive, real-time fitting on a laboratory workstation with a graphical interface. Examples of fits to experimental data will be given, including multicomponent CW EPR spectra as well as two- and three-dimensional FT EPR spectra. Emphasis is placed on the analytic information available from the partial derivatives utilized in the algorithm, and how it may be used to estimate the condition and uniqueness of the fit, as well as to estimate confidence limits for the parameters in certain cases.
|
|
|
Millimeter Wave Electron Spin Resonance Using Quasioptical Techniques
K.A. Earle, D.E. Budil, and J.H. Freed.
Adv. Magn. Optical Reson. 19, 253-323 (1996).
<doi: 10.1016/S1057-2732(96)80019-2>
Publication #231
|
|
|
Summary: We have presented a complete analysis of the Cornell mm-wave spectrometer using quasioptical formalism with sufficient flexibility to predict the performance of a novel reflection spectrometer with variable input coupling and transmit-receive duplexing based on polarization coding and we present a practical realization of these design concepts in Earle et al. (1996b). At every stage we have chosen parameters that correspond to practical performance values and measured response.
The treatment given here is self-contained, but the references contain many useful extensions of our results and alternative treatments may deepen the reader's understanding. We have tried, where possible, to take advantage of EPR spectroscopists' knowledge of microwave circuits and analysis. As high-field ESR becomes more common, we predict that analogies from other fields will continue to be a useful method for extending the generality and utility of the method. Implementing the advanced techniques discussed in Sections IX-XI will give significant inprovements in signal-to-noise ratio as we have demonstrated elsewhere (Earle et al., 1996b).
|
|
|
Rotational Dynamics of Axially Symmetric Solutes in Isotropic Solvents. II. The Stochastic Model
A. Polimeno, G.J. Moro, and J.H. Freed.
J. Chem. Phys. 104, 1090-1104 (1996).
<doi: 10.1063/1.470764>
Publication #230
|
|
|
ABSTRACT: In paper I of this series, a molecular dynamics (MD) study of liquid chlorine was performed, and it includes the definition and observation of operational cage variables. These cage variables were used to describe the local environment of a rotating axially symmetric molecule, or probe. Probe and cage properties of interest, such as rotational correlation functions and momentum correlation functions, were computed, together with an effective distribution of librational cage frequencies. In the second part of this work, we develop a stochastic model which includes the relevant elementary relaxation processes previously identified by the MD study. This stochastic model is based upon a multi-dimensional Fokker–Planck equation for the coupled dynamics of the probe and cage orientations, the angular momentum of the probe, and the librational frequencies for the probe in the cage. Semi-analytical approximations, based upon a "Born–Oppenheimer"-type separability of fast and slow variables, are used in order to calculate probe and cage correlation functions, and they are found to be in reasonable agreement with the MD results. In an appendix the Born–Oppenheimer approximation for stochastic operators is developed.
|
|
|
Rotational Dynamics of Axially Symmetric Solutes in Isotropic Liquids. I. A Collective Cage Description From Molecular Dynamics Simulations
A. Polimeno, G.J. Moro, and J.H. Freed.
J. Chem. Phys. 102, 8094-8106 (1995).
<doi: 10.1063/1.469220>
Publication #229
|
|
|
ABSTRACT: An operational definition of collective cage variables previously introduced for liquid argon is extended, via a molecular dynamics study, to the rotational properties of axially symmetric molecules. Quantitative measures of the static and dynamic cage properties are extracted for liquid Cl2 near the triple point. The collective cage variables are well described by the potential acting on an arbitrary molecule (i.e. solute) for a fixed configuration of the other molecules (i.e. solvent). A dynamic separability of the solute orientation relative to the cage potential and of the relative solute displacement is justified in part by the faster relaxation found for the latter. Large and persistent orientational cage potentials (˜15–20 kBT) lead to substantial alignment of the solute in the cage with an average local order parameter of 0.87. The reorientational correlation times for the cage are consistent with axially symmetric Brownian motion. The reorientational correlation times for the solute are nearly equal to the equivalent ones of the cage, consistent with the strong coupling of solute within its cage which leads to a collective reorientation of solute and cage (e.g. τcage(2)=1.4 ps, and τsolute(2)=1.2 ps). Solute librations within the cage are much faster (τlibr(2)=0.12 ps) and are comparable to the relaxation of the relative solute displacements (τr=0.15 ps). The solute angular momentum exhibits the fastest correlation time (τJ=0.06 ps). While the orientational cage potential shows rapidly and slowly relaxing components (τωf=0.14 ps and τωs=2.87 ps, respectively), its dominant portion shows a very long persistence.
|
|
|
Nuclear modulation effects in "2+1" electron spin-echo correlation spectroscopy
A. Raitsimring, R.H. Crepeau, and J.H. Freed.
J. Chem Phys. 102, 8746-8762 (1995).
<doi: 10.1063/1.468978>
Publication #228
|
|
|
ABSTRACT: This work represents the synthesis of the 2+1 pulse sequence for the study of electron spin-echo envelope modulation (ESEEM) with the technique of spin-echo correlated spectroscopy (SECSY), which has previously been used to study nuclear modulation by two-dimensional Fourier transform ESR methods. This example of "pulse adjustable" spectroscopy, wherein the pulse width and pulse amplitude of the second pulse in a three pulse sequence are introduced as adjustable parameters, leads to enhanced resolution to the key features of the nuclear modulation that are important for structural studies. This is demonstrated in studies on (i) a single crystal of irradiated malonic acid and (ii) a frozen solution of diphenylpicrylhydrazyl in toluene. In particular, it is shown for (i) how the nuclear modulation cross peaks can be preferentially enhanced relative to the autopeaks and to the matrix proton peaks, and also how the autopeaks can be significantly suppressed to enhance resolution for low-frequency cross peaks. For (ii) the low-frequency 14N nuclear modulation could be suppressed leaving just the high-frequency matrix 1H modulation. Additionally, the T2 homogeneous linewidth broadening in the f1 frequency direction is removed in 2+1 SECSY. These features significantly improve resolution to the modulation decay, which is the main observable utilized for distance measurements by ESEEM, compared to SECSY. A simple example of a distance measurement to matrix protons is presented for (ii). It is shown that a major advantage of the 2D format is that the full spin-echo shape is collected, which permits one to study how the effect of the nuclear modulation upon the echo varies with evolution time t1 after the first pulse, and thereby to detect the important modulation features. Additionally it allows for correlating the modulation cross peaks in making spectral assignments. A detailed quantitative theory for 2+1 SECSY is also presented. In general, very good agreement between experiment and theoretical simulations is obtained.
|
|
|
Quenching of the Fluorescence from Chromium(III) Ions in Chromium-Doped Forsterite by an Aluminum Codopant
J.L. Mass, J.M. Burlitch, D.E. Budil, J.H. Freed, D.B. Barber, C.R. Pollock, M. Hischi, and R. Dieckmann.
Chem. Materials 7, 1008-1014 (1995).
<doi: 10.1021/cm00053a030>
Publication #227
|
|
|
ABSTRACT: The H202-assisted sol–gel method was used to synthesize polycrystalline chromium and aluminum codoped forsterite powders with varying ratios of aluminum to chromium. These powders were used as feedstocks to grow single crystals by the floating zone method. The dopant
composition of the resulting pleochroic crystals was determined by neutron activation analysis. Hyperfine splittings observed in the EPR study revealed that over half of the Cr3+ ions were present as chromium-aluminum pairs. The near-IR emission spectra of the crystals displayed the high ratios of Cr4+ to Cr3+ fluorescence intensities that have been previously reported for co-doped forsterite. Integration of the Cr4+ and Cr3+ EPR resonances, however, revealed that most of the chromium was in the 3+ oxidation state. The high ratio of Cr4+/Cr3+ emissions is attributed to quenching of the fluorescence from Cr3+
|
|
|
ESR Studies of Molecular Dynamics of A Liquid Crystalline Polyether
D. Xu, J.K. Moscicki, D.E. Budil, J.H. Freed, E. Hall, and C.K. Ober.
Polymer Preprints 35, 809-810 (1994).
Publication #226
|
|
|
INTRODUCTION: Fundamental knowledge of macroscopic chain dynamics within a polymer melt is essential for a clear understanding of molding properties, adhesion and mechanical properties of engineering resins. A model liquid crystalline (LC) polymer system was developed and its melt diffusion was studied by Forward Recoil Spectrometry (FRES) for diffusion slower than 10−10 cm2∕s. Recently, a new technique called Dynamic Imaging of Diffusion by Electron Spin Resonance (DID-ESR) has been developed to measure translational diffusion of LC in the range of 10−5 to 10−8 cm2∕s with much success. The material must be labeled with a paramagnetic species, usually a stable free radical. As a microscopic technique, ESR is effective in studying the conformation and dynamic molecular structure. DID-ESR combines the precision of microscopic techniques with the long-time scales (for Fickian diffusion) of mass transport techniques. Its primary advantage is its ability to measure rotational diffusion coefficients and order parameters on the same sample that is used for translational diffusion measurements. In this way, macroscopic and microscopic properties of a sample can be related to an unprecedented degree.
The investigation reported here is the initial work to adapt the DID-ESR technique to study short chain LC polymers. The same model polymer system used in previous FRES studies was spin labeled. This publication focuses on the behavior of low molecular weight LC polymers for which liquid crystalline behavior may be important. Melt diffusion coefficients are measured for samples with degrees of polymerization ranging from 12 ˜ 66.
|
|
|
Theory of Two-Dimensional Fourier Transform Electron Spin Resonance for Ordered and Viscous Fluids
S. Lee, D.E. Budil, and J.H. Freed.
J. Chem. Phys. 101, 5529-5558 (1994).
<doi: 10.1063/1.467342>
Publication #225
|
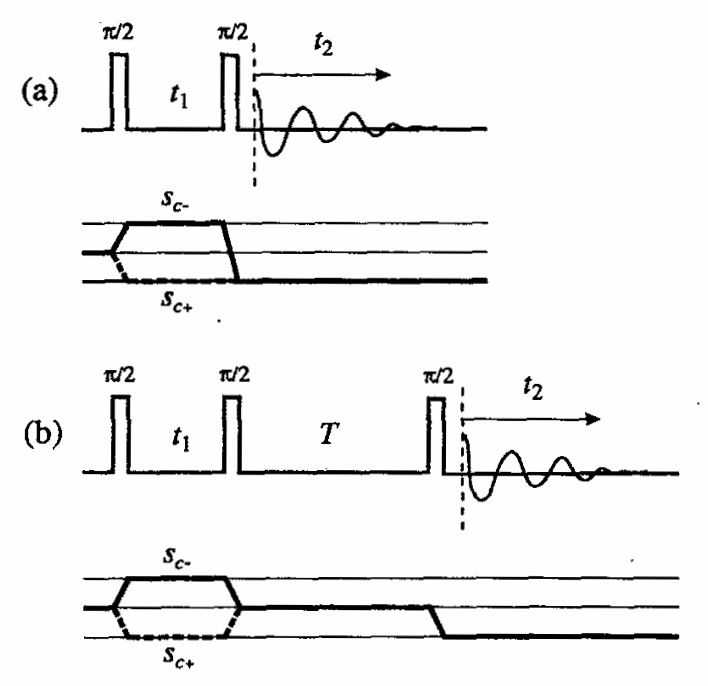
|
|
ABSTRACT: A comprehensive theory for interpreting two-dimensional Fourier transform (2D-FT) electron spin resonance (ESR) experiments that is based on the stochastic Liouville equation is presented. It encompasses the full range of motional rates from fast through very slow motions, and it also provides for microscopic as well as macroscopic molecular ordering. In these respects it is as sophisticated in its treatment of molecular dynamics as the theory currently employed for analyzing cw ESR spectra. The general properties of the pulse propagator superoperator, which describes the microwave pulses in Liouville space, are analyzed in terms of the coherence transfer pathways appropriate for COSY (correlation spectroscopy), SECSY (spin–echo correlation spectroscopy), and 2D-ELDOR (electron–electron double resonance) sequences wherein either the free-induction decay (FID) or echo decay is sampled. Important distinctions are made among the sources of inhomogeneous broadening, which include (a) incomplete spectral averaging in the slow-motional regime, (b) unresolved superhyperfine structure and related sources, and (c) microscopic molecular ordering but macroscopic disorder (MOMD). The differing effects these sources of inhomogeneous broadening have on the two mirror image coherence pathways observed in the dual quadrature 2D experiments, as well as on the auto vs crosspeaks of 2D-ELDOR, is described. The theory is applied to simulate experiments of nitroxide spin labels in complex fluids such as membrane vesicles, where the MOMD model applies and these distinctions are particularly relevant, in order to extract dynamic and ordering parameters. The recovery of homogeneous linewidths from FID-based COSY experiments on complex fluids with significant inhomogeneous broadening is also described. The theory is applied to the ultraslow motional regime, and a simple method is developed to determine rotational rates from the broadening of the autopeaks of the 2D-ELDOR spectra as a function of the mixing time, which is due to the development of "motional crosspeaks." The application of this method to recent experiments with nitroxide probes illustrates that rotational correlation times as slow as milliseconds may be measured. It is shown how 2D-ELDOR can be useful to distinguish between the cases of very slow motional (SM) rates with little or no ordering and of very high ordering (HO) but substantial motional rates even though the cw ESR spectra are virtually the same. The effects of motion and of microscopic ordering on the nuclear modulation patterns in 2D-FT-ESR are compared, and it is suggested that these effects could be utilized to further distinguish between SM and HO cases. Key aspects of the challenging computational problems are discussed, and algorithms are described which lead to significant reductions in computation time as needed to permit nonlinear least-squares fitting of the theory to experiments.
|
|
|
9.6 GHz and 34 GHz Electron Paramagnetic Resonance Studies of Chromium-Doped Forsterite
D.E. Budil, D.G. Park, J.M. Burlitch, R.F. Geray, R. Dieckmann, and J.H. Freed.
J. Chem. Phys. 101, 3538-3548 (1994).
<doi: 10.1063/1.467540>
Publication #224
|
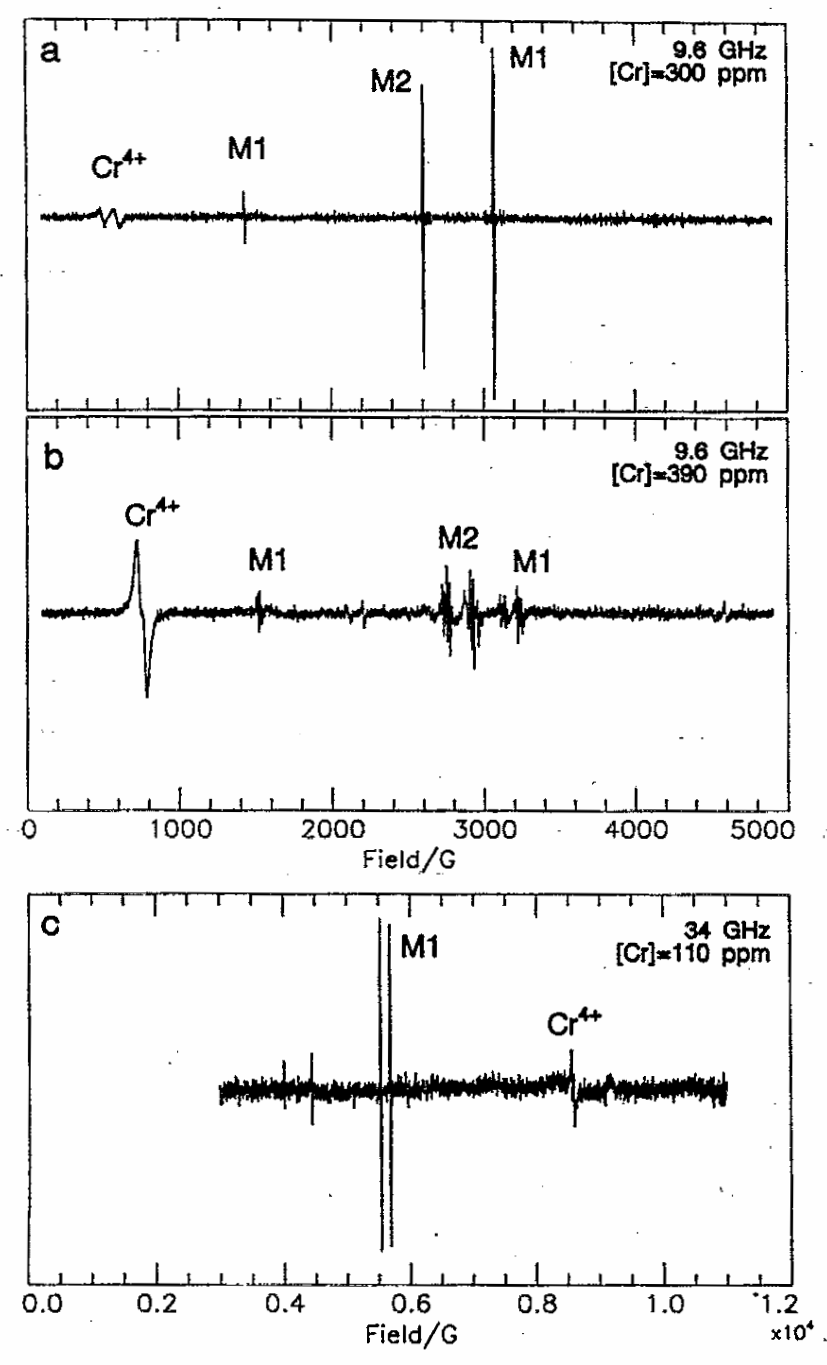
|
|
ABSTRACT: Chromium-doped forsterite single crystals grown under conditions that produce a high Cr4+∕Cr3+ ratio were examined by electron paramagnetic resonance (EPR) at 9.6 and 34 GHz. The crystals were grown in 2–3 atm of oxygen by the floating-zone method starting from polycrystalline chromium-doped forsterite powder synthesized via a sol–gel method. Three crystals with chromium concentrations of 110, 300, and 390 ppm were studied. At 34 GHz, transitions are observed for the laser-active tetrahedral Cr4+ species that are not observable at 9.6 GHz, which improve the resolution and accuracy with which the magnetic parameters can be measured by EPR. In addition, peaks for a non-Kramers species appear at 34 GHz that were not observed at 9.6 GHz. These peaks are not analyzed in detail, but are tentatively ascribed to Cr4+ in the octahedral substitution sites of the crystal. At the highest chromium concentration, the Cr3+ spectra show evidence of direct interaction with Cr4+. A global least-squares fit of the combined 9.6 and 34 GHz data for the 300 ppm crystal gives D=64.26±0.18 GHz, E=−4.619±0.009 GHz, gx=1.955±0.009, gy=2.005±0.040, gz=1.965±0.006, and places the magnetic z axis in the ab plane at an angle of 43.8±0.3° from the b crystallographic axis (in Pbnm). A method for accurately measuring the Cr4+∕Cr4+ ratio using EPR line intensities is given. The EPR linewidth of the Cr4+ center exhibits a strong orientation dependence that is well-modeled by including site variations in the D and E zero-field splittings and in the orientation of the z magnetic axis. The linewidth analysis reveals a high degree of correlation between the distributions in D and E, and a somewhat weaker correlation between E and the z axis orientation. These results are interpreted to suggest that the tetrahedral Cr4+ sites vary mainly in the degree of compression of the tetrahedral cage along the a crystallographic axis. The Cr4+ EPR linewidths increase significantly at higher chromium concentration, but maintain the same qualitative orientation dependence. The EPR data indicate that the major contribution to inhomogeneity in the tetrahedral site, which may be related to the tunable range of the Cr4+ laser center, is distortion induced by chromium substitution into the crystal lattice rather than direct chromium–chromium interactions.
|
|
|
ESR Studies of Spin-Labeled Membranes Aligned by Isopotential Spin-Dry Ultracentrifugation: Lipid-Protein Interactions
M. Ge, D.E. Budil, and J.H. Freed.
Biophys. J. 67, 2326-2344 (1994).
<doi: 10.1016/S0006-3495(94)80719-2>
PMID:
7535112
PMCID:
PMC1225617
Publication #223
|
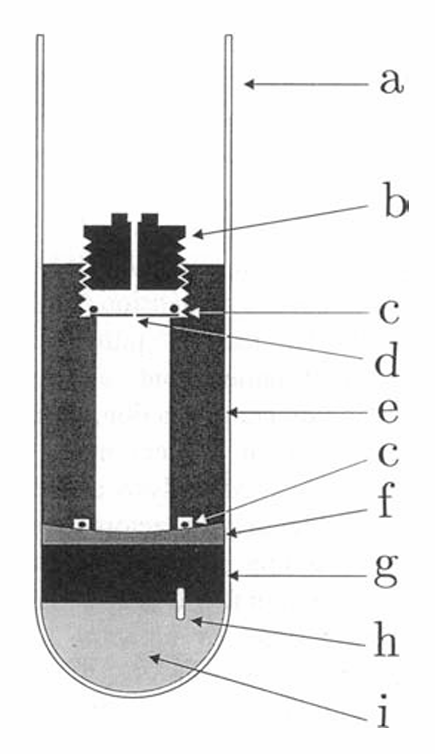
|
|
ABSTRACT: Electron spin resonance (ESR) studies have been performed on spin-labeled model membranes aligned using the isopotential spin-dry ultracentrifugation (ISDU) method of Clark and Rothschild. This method relies on sedimentation of the membrane fragments onto a gravitational isopotential surface with simultaneous evaporation of the solvent in a vacuum ultracentrifuge to promote alignment. The degree of alignment obtainable using ISDU, as monitored by ESR measurements of molecular ordering for both lipid (16-PC) and cholestane spin labels (CSL), in dipalmitoylphosphatidylcholine (DPPC) model membranes compares favorably with that obtainable by pressure-annealing. The much gentler conditions under which membranes may be aligned by ISDU greatly extends the range of macroscopically aligned membrane samples that may be investigated by ESR. We report the first ESR study of an integral membrane protein, bacteriorhodopsin (BR) in well-aligned multilayers. We have also examined ISDU-aligned DPPC multilayers incorporating a short peptide gramicidin A′ (GA), with higher water content than previously studied. 0.24 mol % BR∕DPPC membranes with CSL probe show two distinct components, primarily in the gel phase, which can be attributed to bulk and boundary regions ofthe bilayer. The boundary regions show sharply decreased molecular ordering and spectral effects comparable to those observed from 2 mol % GA∕DPPC membranes. The boundary regions for both BR and GA also exhibit increased fluidity as monitored by the rotational diffusion rates. The high water content of the GA∕DPPC membranes reduces the disordering effect as evidenced by the reduced populations of the disordered components. The ESR spectra obtained slightly below the main phase transition of DPPC from both the peptide- and protein-containing membranes reveals a new component with increased ordering of the lipids associated with the peptide or protein. This increase coincides with a broad endothermic peak in the DSC, suggesting a disaggregation of both the peptide and the protein before the main phase transition of the lipid. Detailed simulations of the multicomponent ESR spectra have been performed by the latest nonlinear least-squares methods, which have helped to clarify the spectral interpretations. It is found that the simulations of ESR spectra from CSL in the gel phase for all the lipid membranes studied could be significantly improved by utilizing a model with CSL molecules existing as both hydrogen-bonded to the bilayer interface and non-hydrogen-bonded within the bilayer.
|
|
|
Two-Dimensional Fourier-Transform Electron Spin Resonance in Complex Fluids
S. Lee, B.R. Patyal, S. Saxena, R.H. Crepeau, and J.H. Freed.
Chem. Phys. Letts. 221, 397-406 (1994).
<doi: 10.1016/0009-2614(94)00281-9>
Publication #222
|
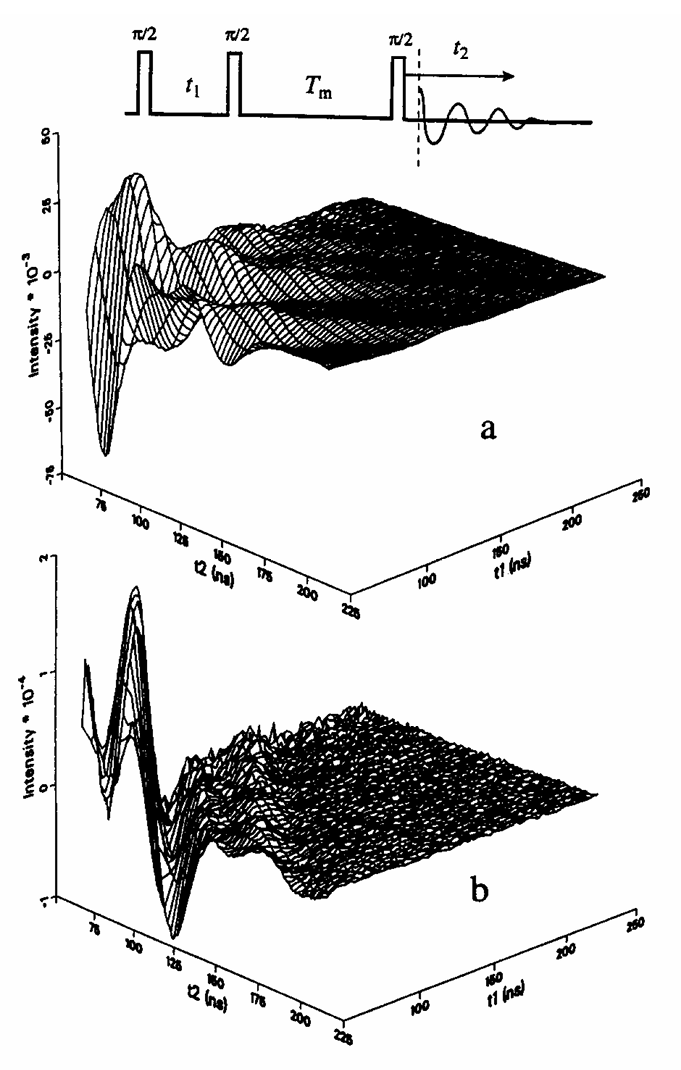
|
|
ABSTRACT: Two-dimensional Fourier-transform ESR spectra from a complex fluid characterized by very short free induction decay times are reported. They provide enhanced resolution to dynamic molecular structure compared to conventional cw-ESR spectra which are inhomogeneously broadened by the macroscopic disorder. A general theoretical analysis based on the stochastic Liouville equation permits us to accurately determine the microscopic ordering and motional dynamic rates by non-linear least-squares fitting to experiment.
|
|
|
250-GHz Electron Spin Resonance Studies of Polarity Gradients Along the Aliphatic Chains in Phospholipid Membranes
K.A. Earle, J.K. Moscicki, M. Ge, D.E. Budil, and J.H. Freed.
Biophys. J. 66, 1213-1221 (1994).
<doi: 10.1016/S0006-3495(94)80905-1>
PMID:
7518705
PMCID:
PMC1275829
Publication #221
|
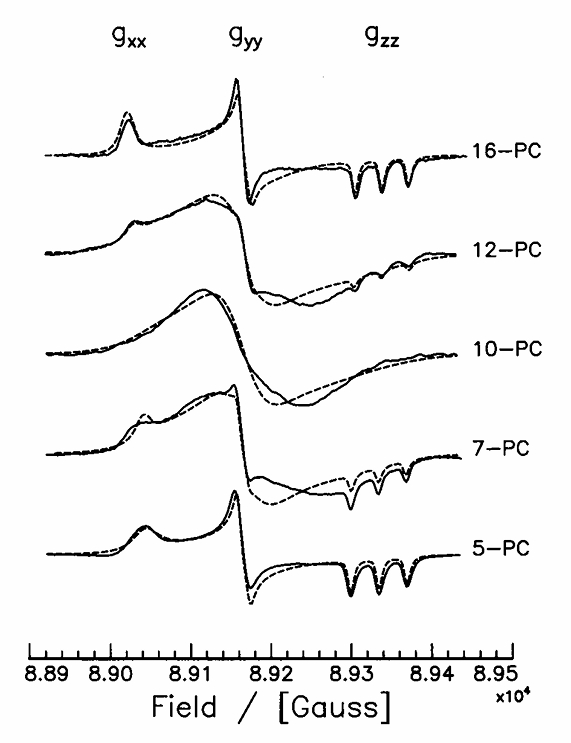
|
|
ABSTRACT: Rigid-limit 250-GHz electron spin resonance (FIR-ESR) spectra have been studied for a series of phosphatidylcholine spin labels (n-PC, where n = 5, 7, 10, 12, 16) in pure lipid dispersions of dipalmitoylphosphatidylcholine (DPPC) and 1-palmitoyl-2-oleoylphosphatidylcholine (POPC), as well as dispersions of DPPC containing the peptide gramicidin A (GA) in a 1:1 molar ratio. The enhanced g-tensor resolution of 250-GHz ESR for these spin labels permitted a careful study of the nitroxide g-tensor as a function of spin probe location and membrane composition. In particular, as the spin label is displaced from the polar head group, Azz decreases and gxx increases as they assume values typical of a nonpolar environment, appropriate for the hydrophobic alkyl chains in the case of pure lipid dispersions. The field shifts of spectral features due to changes in gxx are an order of magnitude larger than those from changes in Azz. The magnetic tensor parameters measured in the presence of GA were characteristic of a polar environment and showed only a very weak dependence of Azz and gxx on label position. These results demonstrate the significant influence of GA on the local polarity along the lipid molecule, and may reflect increased penetration of water into the alkyl chain region of the lipid in the presence of GA. The spectra from the pure lipid dispersions also exhibit a broad background signal that is most significant for 7-, 10-, and 12-PC, and is more pronounced in DPPC than in POPC. It is attributed to spin probe aggregation yielding spin exchange narrowing. The addition of GA to DPPC essentially suppressed the broad background signal observed in pure DPPC dispersions.
|
|
|
An Electron Spin Resonance Study of Interactions Between Phosphatidylcholine and Phosphatidylserine in Oriented Membranes
M. Ge, D.E. Budil, and J.H. Freed.
Biophys. J. 66, 1515-1521 (1994).
<doi: 10.1016/S0006-3495(94)80942-7>
PMID:
8061200
PMCID:
PMC1275871
Publication #220
|
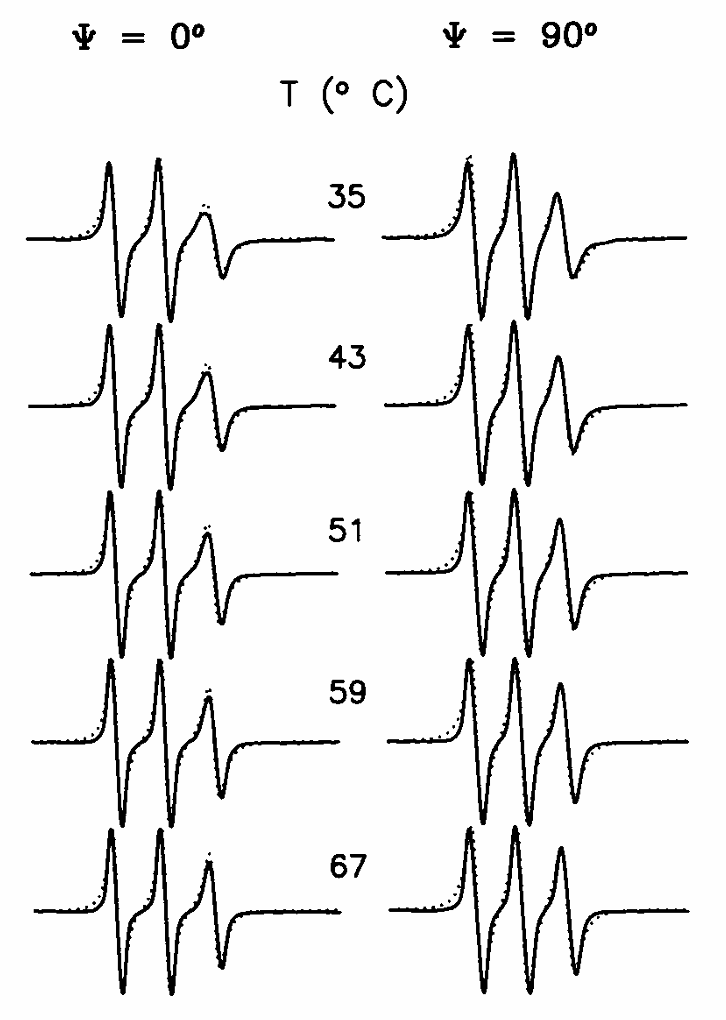
|
|
ABSTRACT: A detailed electron spin resonance (ESR) study of mixtures of 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC) and phosphatidylserine (POPS) in oriented multilayers in the liquid crystalline phase is reported with the purpose of characterizing the effects of headgroup mixing on the structural and dynamical properties of the acyl chains. These studies were performed over a range of blends of POPC and POPS and temperatures, utilizing the spin-labeled lipids 16-phosphatidylcholine and 5-phosphatidylcholine as well as cholestane (CSL). The ESR spectra were analyzed by nonlinear least-squares fitting using detailed spectral simulations. Whereas CSL shows almost no variation in ordering and rotational dynamics versus mole fraction POPS, (i.e. XPS), and 5-PC shows small effects, the weakly ordered end-chain labeled 16-PC shows large relative effects, such that the orientational order parameter, S is at a minimum for XPS = 0.5 where it is about one-third the value observed for XPS = 0 and 1. This is directly reflected in the ESR spectrum as a substantial variation in the hyperfine splitting with XPS. The least-squares analysis also shows a reduction in rotational diffusion coefficient, R⊥ by a fractor of 2 for XPS = 0.5 and permits the estimation of S2, the ordering parameter representing deviations from cylindrically symmetric alignment. These results are contrasted with 2H NMR studies which were insensitive to effects of mixing headgroups on the acyl chains. The ESR results are consistent with a somewhat increased disorder in the end-chain region as well as a small amount of chain tilting upon mixing POPC and POPS. They demonstrate the high sensitivity of ESR to subtle effects in chain ordering and dynamics.
|
|
|
Studies on Lipid Membranes by Two-Dimensional Fourier Transform ESR: Enhancement of Resolution to Ordering and Dynamics
R.H. Crepeau, S. Saxena, S. Lee, B. Patyal, and J.H. Freed.
Biophys. J. 66, 1489-1504 (1994).
<doi: 10.1016/S0006-3495(94)80940-3>
PMID:
8061198
PMCID:
PMC1275869
Publication #219
|
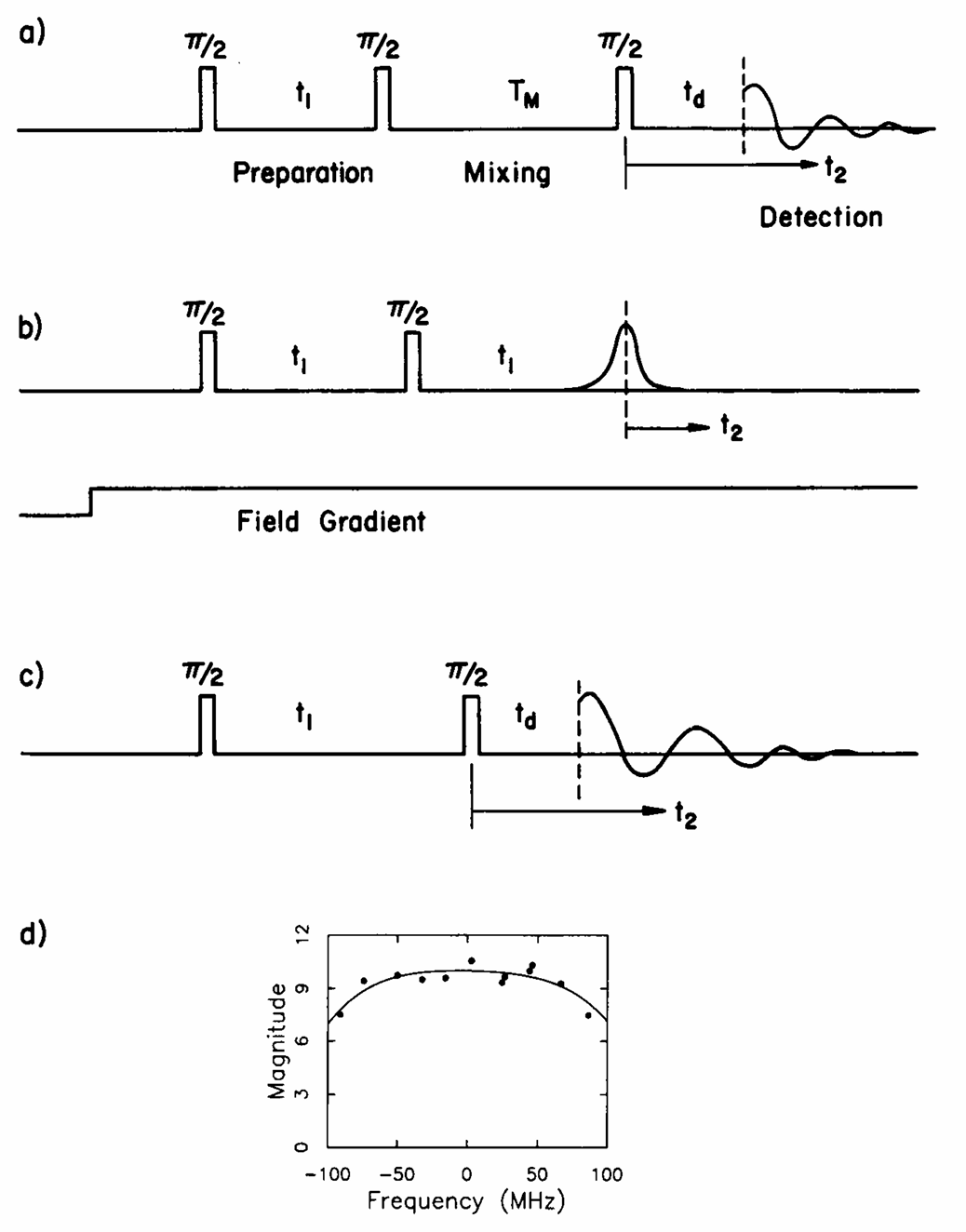
|
|
ABSTRACT: The first two-dimensional Fourier-transform electron spin resonance (2D-FT-ESR) studies of nitroxide-labeled lipids in membrane vesicles are reported. The considerable enhancement this experiment provides for extracting rotational and translational diffusion rates, as well as orientational ordering parameters by means of ESR spectroscopy, is demonstrated. The 2D spectral analysis is achieved using theoretical simulations that are fit to experiments by an efficient and automated nonlinear least squares approach. These methods are applied to dispersions of 1-palmitoyl-2oleoyl-sn-glycerophosphatidylcholine (POPC) model membranes utilizing spin labels 1-palmitoyl-2-(16-doxyl stearoyl) phosphatidylcholine and the 3-doxyl derivative of cholestan-3-one (CSL). Generally favorable agreement is obtained between the results obtained by 2D-FT-ESR on vesicles with the previous results on similar systems studied by continuous wave (cw) ESR on aligned samples. The precision in determining the dynamic and ordering parameters is significantly better for 2D-FT-ESR, even though the cw ESR spectra from membrane vesicles are resolved more poorly than those from well aligned samples. Some small differences in results by the two methods are discussed in terms of limitations of the methods and∕or theoretical models, as well as possible differences between dynamic molecular structure in vesicles versus aligned membranes. An interesting observation with CSL∕POPC, that the apparent homogeneous linewidths seem to increase in "real time," is tentatively attributed to the effects of slow director fluctuations in the membrane vesicles.
|
|
|
250-GHz EPR of Nitroxides in the Slow-Motional Regime: Models of Rotational Diffusion
K.A. Earle, D.E. Budil, and J.H. Freed.
J. Phys. Chem. 97, 13289-13297 (1993).
<doi: 10.1021/j100152a037>
Publication #218
|
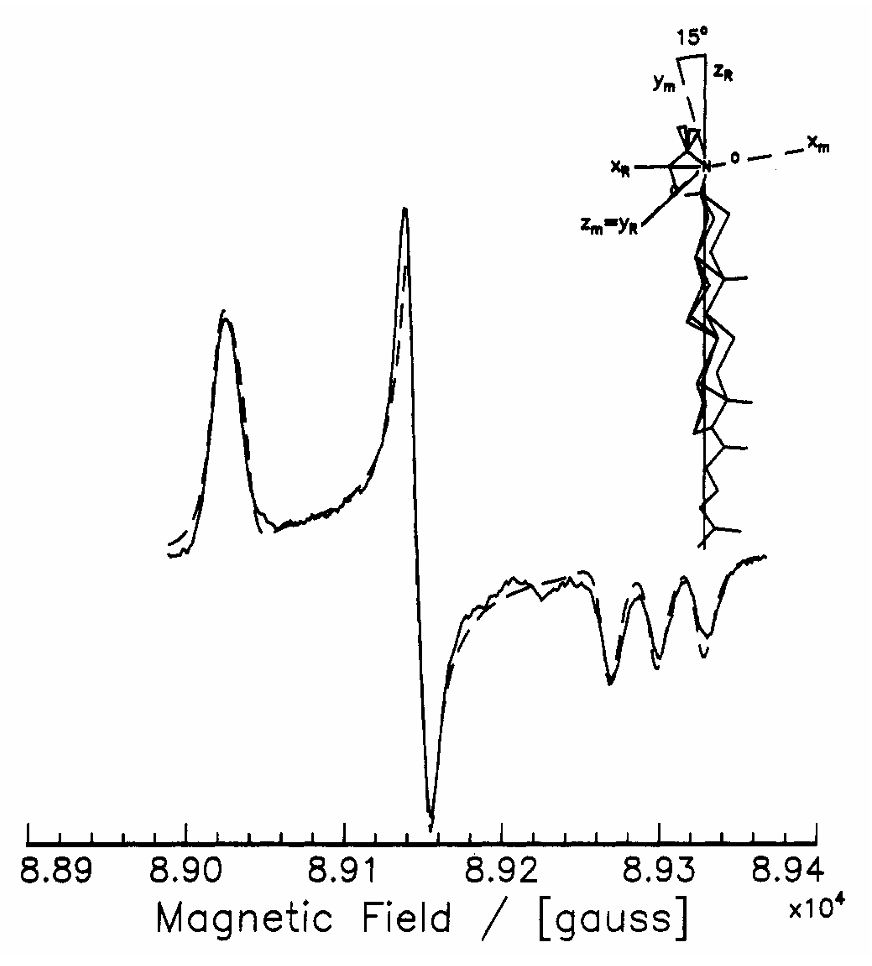
|
|
ABSTRACT: A 250-GHz electron paramagnetic resonance (EPR) study of the slow rotational diffusion of two spin probes in toluene, viz., perdeuterated 2,2,6,6-tetramethyl-4-piperidone (PDT) and 3-doxylcholestane (CSL) is presented. EPR spectra were obtained in the slow-motional and near-rigid limit regions, which corresponds to rotational correlation times 10−10 > τR > 10−6 s. These two probes differ significantly in size and shape, permitting a detailed exploration of the sensitivity of 250-GHz EPR to different aspects of the molecular dynamics such as rotational anisotropy and non-Brownian diffusion. Nonlinear least-squares fitting based on full stochastic Liouville calculations provides a sensitive means for discriminating amongst motional models. PDT in toluene-d8 is found to be well described by an approximate free diffusion model, whereas the larger spin probe, CSL, is best described by Brownian diffusion. The slow-motional spectra at 250 GHz are most sensitive to the diffusional model, the (geometric) mean diffusional rate, and axial diffusional anisotropy but less sensitive to rhombic deviations from an axially symmetric diffusion tensor (i.e., to the general case Rx ≠ Ry ≠ Rz). The slow-motional spectra of PDT were fit using anisotropic diffusion parameters determined from fast-motional spectra but are not very sensitive to such small anisotropies. For the case of Brownian diffusion, CSL was best fit with Ny ≡ Ry∕(RzRx)1∕2 = 9.0 (where the y axis is the long axis of the molecule and x and z are perpendicular axes), which differs appreciably from the fast-motional value of Ny = 4.3 ± 0.2 (and rho;x ≡ Rx∕Rz = 0.5). However, a mixed model of free-diffusional motion about the y axis with Brownian motion of this axis yields an Ny close to the fast-motional value with comparable overall quality in fit compared to full Brownian motion. An important feature of the 250-GHz studies is the ability to measure very accurately the magnetic tensors needed for the motional studies. The theoretical modifications needed for inclusion of a fully anisotropic rotational diffusion tensor in the slow-motional EPR simulations are also given.
|
|
|
Field Gradient ESR and Molecular Diffusion in Model Membranes
J.H. Freed.
Annu. Rev. Biophys. Biomol. Struct. 23, 1-25 1994.
<doi: 10.1146/annurev.bb.23.060194.000245>
PMID:
7919778
Publication #217
|
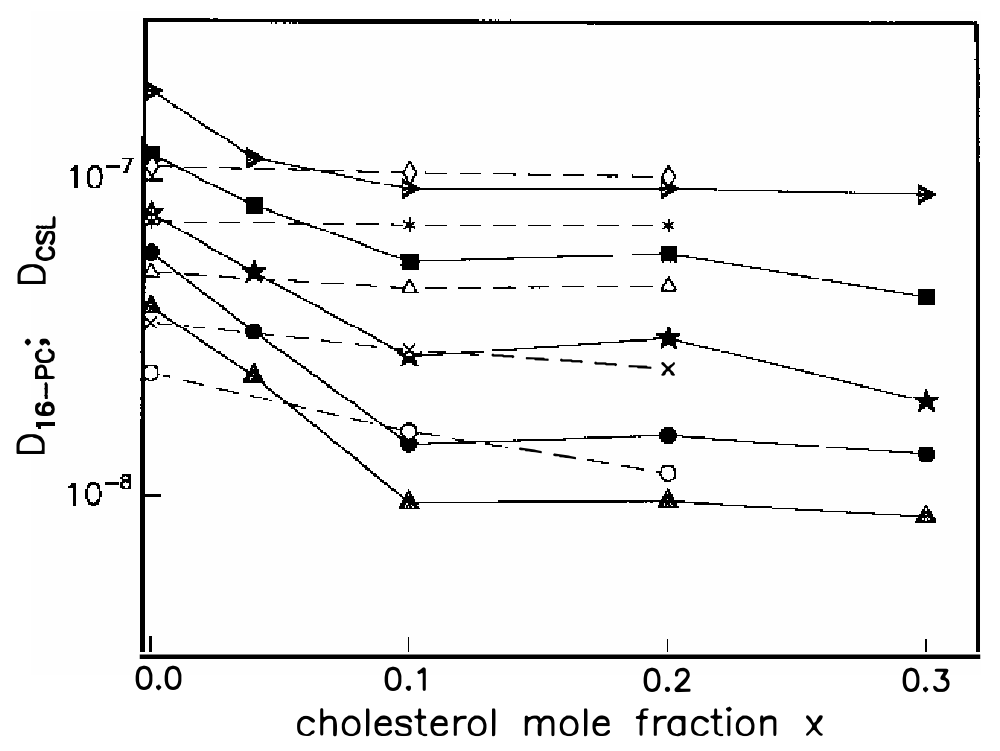
|
|
INTRODUCTION: The dynamic behavior of biomembranes occupies a central focus of modern membrane research. In particular, translational diffusion of lipids and proteins is essential for various biological processes. The techniques developed to measure the translational diffusion coefficients in model or biomembranes can be divided into two general categories according to their relevant distance scales. Modern macroscopic methods include NMR-spin echo, fluorescence recovery after photobleaching (FRAP) and dynamic imaging of diffusion by electron spin resonance (ESR). These enable one to study how the bulk distribution of labeled molecules changes with time and they have resolution on the order of a few to several hundred micrometers. Microscopic methods allow one to observe diffusion over the order of molecular diameters, i.e. a few tens of Ångstroms. Such techniques typically reveal diffusion via encounters between labeled molecules through excimer formation, quasi-elastic neutron scattering, NMR T1 relaxation measurements,
or Heisenberg spin exchange.
|
|
|
A 250-GHz ESR Study of Highly Distorted Manganese Complexes
W.B. Lynch, R.S. Boorse, and J.H. Freed.
J. Am. Chem. Soc. 115, 10909-10915 (1993).
<doi: 10.1021/ja00076a056>
Publication #216
|
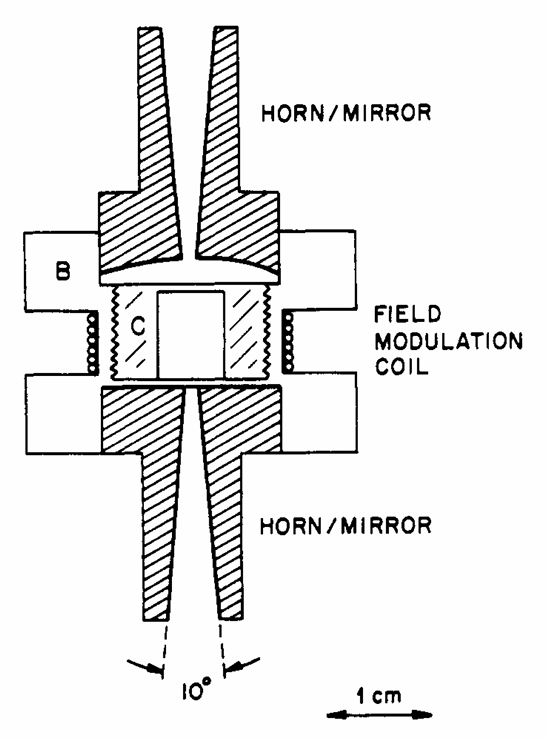
|
|
ABSTRACT: ESR spectra at 250 GHz and magnetic fields ranging from 45 to 95 kG have been measured for the compounds Mn(γ-picoline)4X2 and Mn (o-phenanthroline)2X2, where X = Cl, Br, and I. These spectra are in the high-field limit and well resolved; hence, they are inherently simple to analyze. We find these spectra to be very sensitive to the precise values of the zero-field splitting (zfs) parameters. Thus D and η (E/D) can be estimated directly from the spectrum, with very accurate values obtained using third-order perturbation theory for the electron spin Hamiltonian of high-spin Mn2+. The contributions of all five allowed electron spin transitions are observed and successfully simulated. The γ-picoline complexes have axial symmetry and D increases from 0.186 to 0.999 cm−1 with the size of the halogen. The o-phenanthroline complexes show a wide range of rhombic distortion and E increases linearly with the magnitude of D. The chloride compound is nearly axial (D = 0.124 cm−1, η = 0.04), while the iodide compound is highly distorted (D = 0.590 cm−1, η = 0.246). To demonstrate the applicability of the high-frequency ESR technique to biological samples, we have also measured the high-field spectrum of Mn(II) protoporphyrin IX and determined its zfs parameters to be D = 0.775 cm−1 and η = 0.048.
|
|
|
An Electron Spin Resonance Study of Interactions between Gramicidin A′ and Phosphatidylcholine Bilayers
M. Ge and J.H. Freed.
Biophys. J. 65, 2106-2123 (1993).
<doi: 10.1016/S0006-3495(93)81255-4>
PMID:
7507719
PMCID:
PMC1225946
Publication #215
|
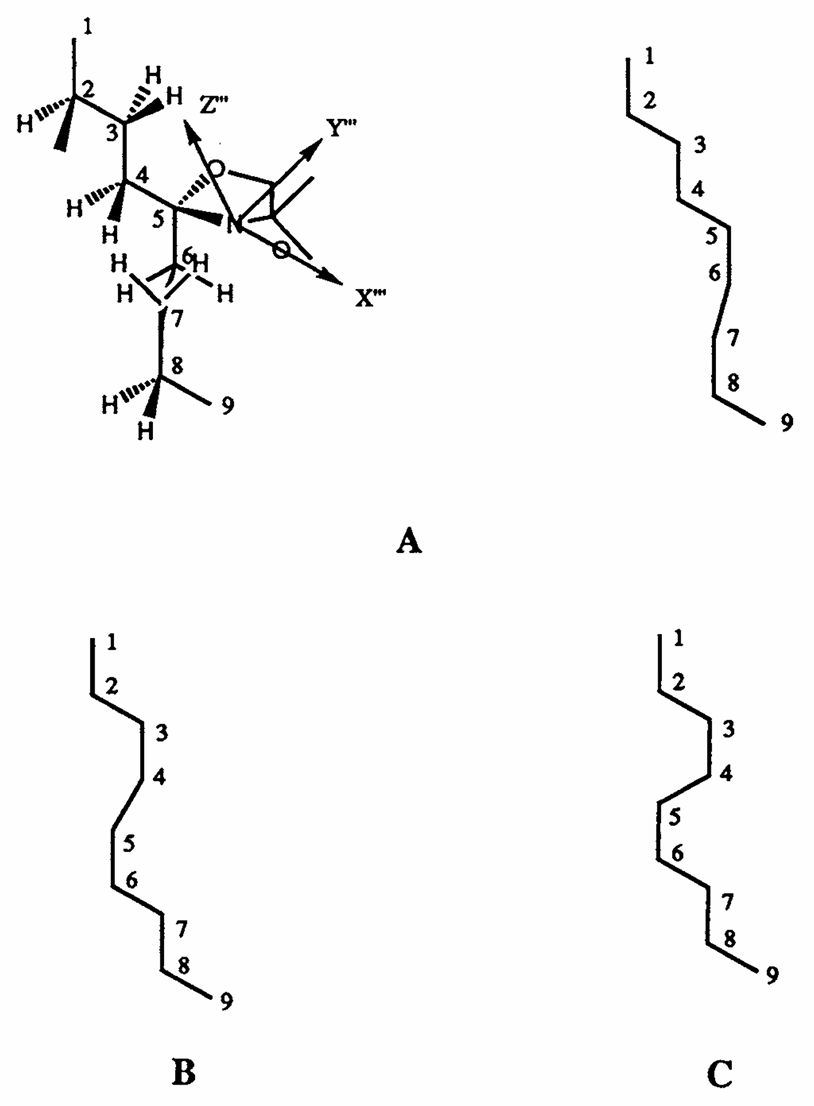
|
|
ABSTRACT: The model of microscopic order and macroscopic disorder was used to stimulate electron spin resonance spectra of spin-labeled lipids, 5-PC, 10-PC, and 16-PC in multilamellar vesicles of dipalmitoylphosphatidylcholine (DPPC) containing gramicidin A′ (GA) at temperatures above the gel-to-liquid crystal transition of DPPC. The simulations show that at a lower concentration of GA (i.e., molar ratios of DPPC∕GA greater than 3), GA has only a slight effect on the acyl chain dynamics. The rotational diffusion rate around the axis parallel to the long hydrocarbon chain remains unchanged or increases slightly, while the rate around the perpendicular axes decreases slightly. These spectra from DPPC∕GA mixtures could only be fit successfully with two or more components consistent with the well-known concept of "boundary lipids," that is, the peptide induces structural inhomogeneity in lipid bilayers. However, the spectra were significantly better fit with additional components that exhibit increased local ordering, implying decreased amplitude of rotational motion, rather than immobilized components with sharply a reduced rotational rate. The largest relative effects occur at the end of the acyl chains, where the average local order parameter S̄t of 16-PC increases from 0.06 for pure lipid to 0.66 for 1:1 DPPC∕GA. The inhomogeneity in ordering in DPPC bilayers due to GA decreases with increasing temperature. The hyperfine tensor component Azz increases for 10-PC and 16-PC when GA is incorporated into DPPC bilayers, indicating that water has deeply penetrated into the DPPC bilayers. Simulations of published electron spin resonance spectra of 14-PC in dimyristoylphosphatidylcholine∕cytochrome oxidase complexes were also better fit by additional components that were more ordered, rather than immobilized. The average local order parameter in this case is found to increase from 0.11 for pure dimyristoylphosphatidylcholine to 0.61 for a lipid∕protein ratio of 50. These spectra and their simulations are similar to the results obtained with 16-PC in the DPPC∕GA mixtures. The relevance to studies of lipid-protein interactions for other proteins is briefly discussed.
|
|
|
Translational Diffusion in a Smectic-A Phase by Electron Spin Resonance Imaging: The Free-Volume Model
J.K. Moscicki, Y.-K. Shin, and J.H. Freed.
J. Chem. Phys. 99, 634-649 (1993).
<doi: 10.1063/1.465736>
Publication #214
|
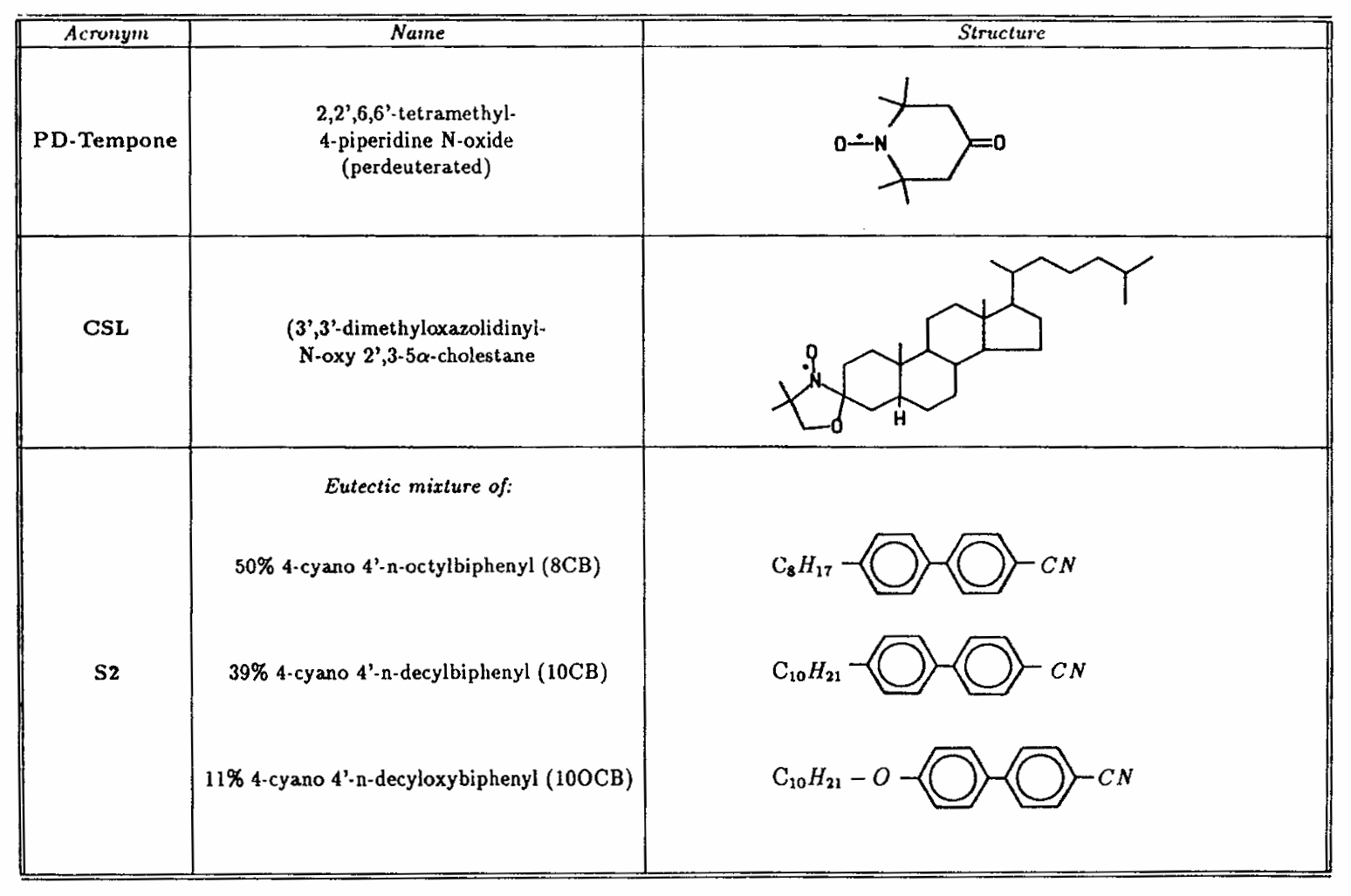
|
|
ABSTRACT: The method of dynamic imaging of diffusion (DID)-ESR (electron spin resonance) has been utilized to study the anisotropy of translational diffusion of spin probes in the smectic A phase of a eutectic liquid crystal, S2. In particular, the nearly spherical perdeuterated-TEMPONE (PDT) and the rigid and elongated cholestane spin label (CSL) molecules were studied. Whereas D⊥ (the coefficient of diffusion perpendicular to the nematic director) showed simple Arrhenius dependence for both probes, diffusion parallel to the director displayed two different temperature regimes with a changeover of D∥ at
t*≈26–27 °C. The regime above (below) t* is characterized by weak (strong) translational ordering. For CSL the ratio D⊥∕D∥<1 above t* which indicates nematiclike behavior, but below t* the behavior is more smecticlike, i.e., D⊥∕D∥>1; for PDT D⊥∕D∥>1 over the whole temperature range. A free volume model is developed to interpret the activation energies associated with D⊥ and D∥ (i.e., E⊥ and E∥) in terms of the orientational and translational order parameters for the smectic phase and those for the spin probes. Also included are the variation of the compressibility across the smectic layer and the length of the probe relative to that of the thickness of the smectic layer. The fact that above t* E⊥∕E∥ is unity for CSL but a little greater that unity for PDT is interpreted as due to the weaker coupling of the larger CSL molecule to the weak translational ordering and compressibility variation. Below t*, E⊥∕E∥ becomes 1.52 and 1.80 for CSL and PDT, respectively, which may be interpreted in terms of enhancement of these smectic features. The free volume model may be used to analyze E∥ and E⊥ for self-diffusion and for a wide range of spin probes, including such very small probes like methane, as a function of the key parameters.
|
|
|
Thermodynamics and Dynamics of Phosphatidylcholine-Cholesterol Mixed Model Membranes in the Liquid Crystalline State: Effects of Water
Y.-Kyun Shin, D.E. Budil, and J.H. Freed.
Biophys. J. 65, 1283-1294 (1993).
<doi: 10.1016/S0006-3495(93)81160-3>
PMID:
8241408
PMCID:
PMC1225848
Publication #213
|
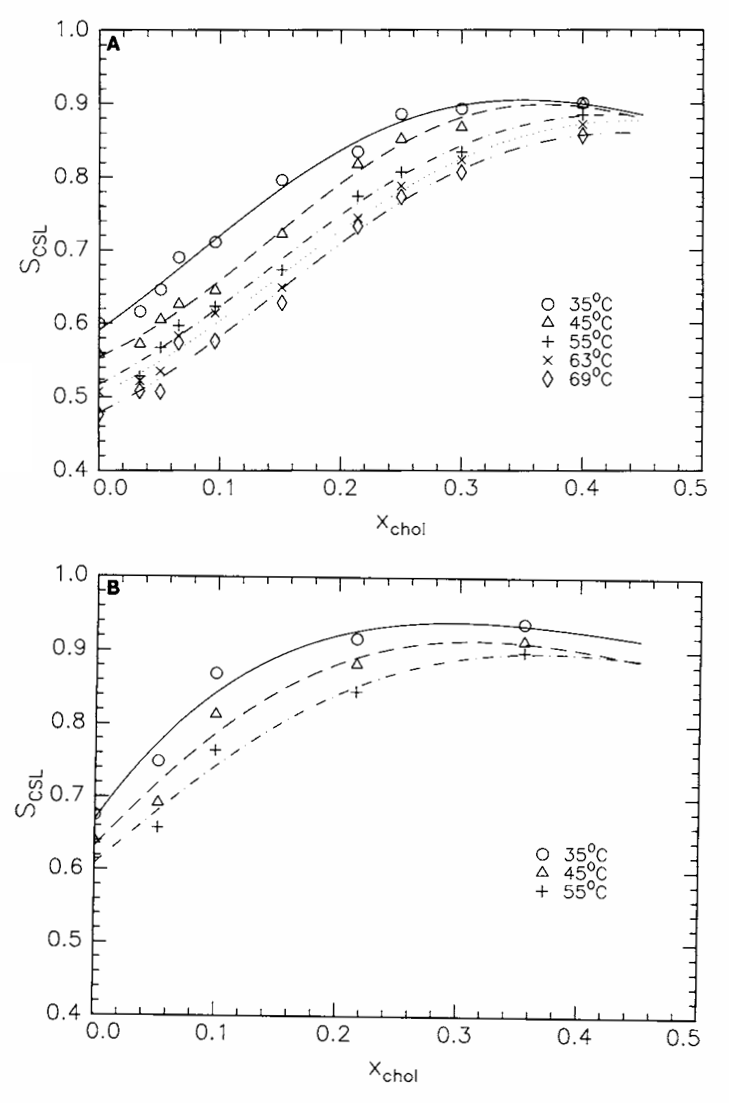
|
|
ABSTRACT: A method for obtaining the thermodynamic activity of each membrane component in phosphatidylcholine (PC)∕cholesterol mixtures, that is based upon ESR spin labeling is examined. The thermodynamic activity coefficients, γPC and γchol, for the PC and cholesterol, respectively, are obtained from the measured orientational order parameters, SPC and Schol, as a function of cholesterol content for a spin-labeled PC and the sterol-type cholestane spin probe (CSL), respectively, and the effects of water concentration are also considered. At water content of 24 weight%, the thermodynamics of DMPC∕cholesterol∕water mixtures in the liquid-crystalline state may be treated as a two-component solution ignoring the water, but at lower water content the role of water is important, especially at lower cholesterol concentrations. At lower water content (17 wt%), γchol decreases with increasing cholesterol content which implies aggregation. However, at higher water content (24 wt%), γchol is found initially to increase as a function of cholesterol content before decreasing at higher cholesterol content. This implies a favorable accommodation for the cholesterol in the membrane at high water and low cholesterol content. Good thermodynamic consistency according to the Gibbs-Duhem equation was obtained for γPC and γchol at 24 wt% water. The availability of γchol (and γPC) as a function of cholesterol concentration permits the estimate of the boundary for phase separation.
The rotational diffusion coefficients of the labeled PC and of CSL were also obtained from the ESR spectra. A previously proposed universal relation for the perpendicular component of the rotational diffusion tensor, Rperp;, for CSL in PC∕cholesterol mixtures (i.e., Rperp; = R0perp; exp(-AS2chol∕ℛT)) is confirmed. A change in composition of cholesterol or of water for DMPC∕cholesterol∕water mixtures affects Rperp; only through the dependence of Schol on the composition. In particular, the amount of water affects the membrane fluidity, monitored by Rperp; for CSL, solely by the structural changes it induces in the membrane for the compositions studied. Rotational diffusion for the labeled PC is found to be more complex, most likely due to the combined action of the internal modes of motion of the flexible chain and of the overall molecular reorientation.
|
|
|
250- and 9.5-GHz EPR Studies of an Electride and Two Alkalides
D.H. Shin, J.L. Dye, D.E. Budil, K.A. Earle, and J.H. Freed.
J. Phys. Chem. 97, 1213-1219 (1993).
<doi: 10.1021/j100108a017>
Publication #212
|
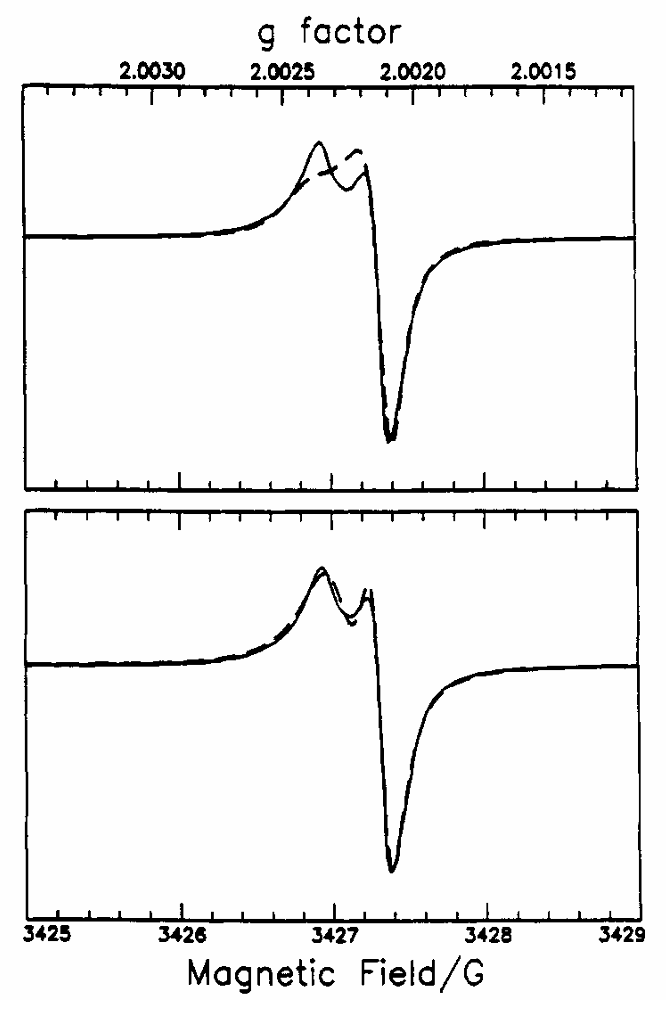
|
|
ABSTRACT: The EPR spectra of polycrystalline samples of Cs+(18-crown-6)2X−, in which X− = e−, Na−, or Cs−, were studied at both X-band (9 GHz) and at 250 GHz. The high frequency affords much better g-factor resolution and reveals at least two types of asymmetric electron sites in each of these compounds. The defect electrons detected in the alkalides Cs+(18C6)2Na− and Cs+(18C6)2Cs−appear to be located at isolated centers in the crystal lattice with the strongest signal originating from electrons trapped at anion vacancies. These species exhibit broadening due to unresolved superhyperfine interactions at the periphery of the trapping site. In contrast, the X-band spectra of the two-electron sites in the electride appear to be exchange-narrowed, indicating that the electron spins have some degree of mobility within the crystal lattice. Comparison of the 9- and 250-GHz spectra of the electride places an upper bound of about 1 × 107 s−1 on the rate of spin exchange. The temperature and saturation behavior of the electride at X-band are consistent with the "F-center" model, in which electrons serve to balance the charge of the complexed cations, with the center of charge of each electron at an anionic site. The strongest signal in Cs+(18C6)2e− has nearly the same g-anisotropy as one of the signals from the isostructural sodide, Cs+(18C6)2Na−, indicating that at least one of the electron-trapping sites detected in the two crystal lattices is the same. The temperature dependence of the two EPR signals from Cs+(18C6)2Cs− at X-band suggests an activated process that populates two types of unpaired electron sites from a common spin-paired precursor.
|
|
|
Full Determination of the Rotational Diffusion Tensor by Electron Paramagnetic Resonance at 250 GHz
D.E. Budil, K.A. Earle, and J.H. Freed.
J. Phys. Chem. 97, 1294-1303 (1993).
<doi: 10.1021/j100109a009>
Publication #211
|
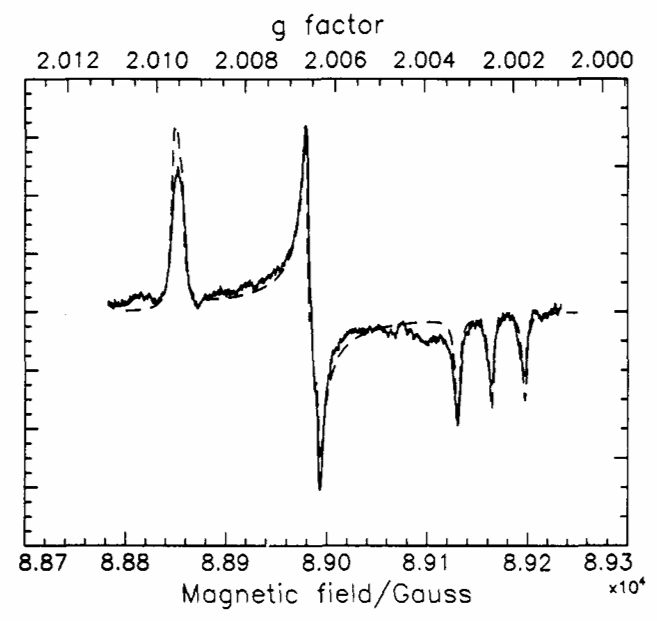
|
|
ABSTRACT: High-frequency (250 GHz) electron paramagnetic resonance (EPR) spectra in the limit of motional narrowing have been studied for a nitroxide spin probe diffusing in a low-viscosity isotropic solvent. The enhanced sensitivity of the 250-GHz spectrum to rotational modulation of the g tensor reveals new details of the microscopic motion of the spin probe. Specifically, it is shown how independent linear constraints imposed by line-width measurements at 250 GHz and a lower frequency may be used to fully determine the diffusion tensor R in the general case Rx ≠ Ry ≠ Rz. This procedure is demonstrated experimentally for the perdeuterated Tempone (PDT) probe diffusing in toluene-d8 where analysis of the line widths obtained at 250 and 9.5 GHz gives values for the anisotropy parameters ρx = Rx∕Rz and ρy = Ry∕Rz of 1.8 ± 0.2 and 1.5 ± 0.3, respectively. Thus, this molecule is found to exhibit small deviations from spherically symmetric reorientation. The enhanced accuracy of determining the g tensor from rigid-limit spectra obtained at 250 GHz is an important feature of such high-frequency studies. Existing theoretical expressions for motionally narrowed nitroxide line widths have been modified appropriately for the high-field case by including a completely anisotropic diffusion tensor and the 14N nuclear Zeeman interaction, which becomes important at fields above about 5 T.
|
|
|
A Two-Dimensional Fourier Transform Electron-Spin Resonance (ESR) Study of Nuclear Modulation and Spin Relaxation in Irradiated Malonic Acid
S. Lee, B.R. Patyal, and J.H. Freed.
J. Chem. Phys. 98, 3665-3689 (1993).
<doi: 10.1063/1.464044>
Publication #210
|
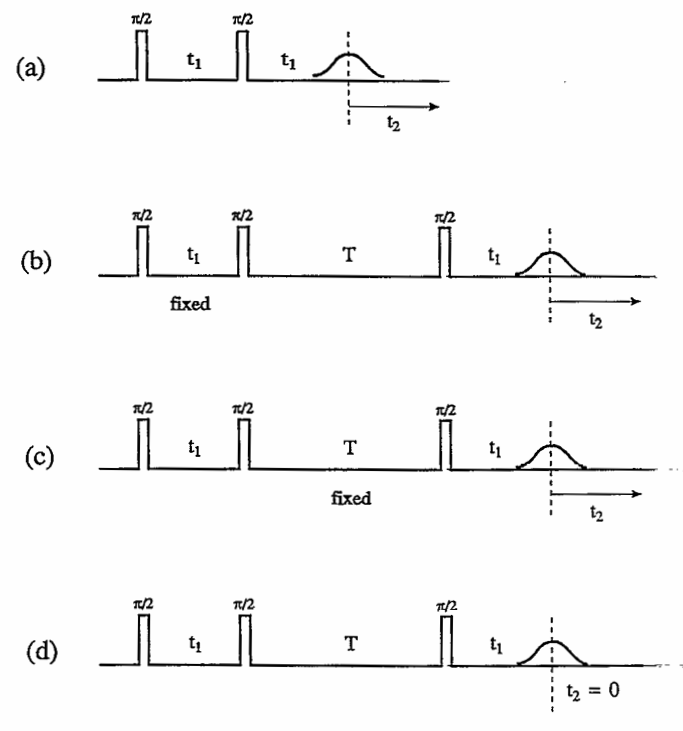
|
|
ABSTRACT: Nuclear modulation in electron-spin-echo spectroscopy is conventionally studied by one-dimensional electron-spin-echo envelope modulation (1D-ESEEM). Two-dimensional Fourier transform electron-spin resonance (2D-FTESR) studies of nuclear modulation have the promise of enhancing the spectral resolution and clarifying the key details of the relaxation processes. We present a 2D-FTESR study on single proton nuclear modulation from γ-irradiated malonic acid single crystals to test the validity of the Gamliel–Freed theory and to assess the value of the new methods. The two pulse spin-echo correlation spectroscopy (SECSY) spectra as a function of orientation of the single crystal show very good agreement with the Gamliel–Freed theory extended to the general case of nonaxially symmetric hyperfine interaction. It is very simply affected by spin relaxation, such that relative intensities are essentially unaffected. Thus SECSY-ESR can most reliably be utilized for studying nuclear modulation. Stimulated SECSY provides the simplest nuclear modulation patterns, which, however, do exhibit the suppression effect well known in three-pulse ESEEM studies. Two-dimensional electron–electron double resonance (2D-ELDOR) provides nuclear modulation patterns similar to that of SECSY-ESR, so the suppression effect is absent. Both three-pulse methods exhibit complex relaxation behavior which can affect relative intensities. This is a feature characteristic of three-pulse ESEEM, but is not well understood. It is shown how the 2D-FTESR methods enable one to obtain the details of the complex spin relaxation, and in the process, obtain very good agreement between experiment and theory. 2D-ELDOR exhibits exchange cross peaks as well as coherence peaks from the nuclear modulation. It is shown how experiments, as a function of mixing time, enable one to separate the effects of the two. It is pointed out that such experiments are in the spirit of 3D spectroscopy. A new observation of the broadening of the 2D-ELDOR main peaks with an increase in mixing time is ascribed to the effects of solid-state dynamical processes that are slow on the ESR time scale and may thus be studied in "real time" in such experiments. The analysis of spin relaxation in this study is enabled by a full Liouville space derivation of the combined effects of nuclear modulation and spin relaxation in 2D-FTESR.
|
|
|
Primary Events in Photosynthesis: Problems, Speculations, Controversies and Future Trends
M. Bixon, J. Fajer, G. Feher, J.H. Freed, D. Gamliel, A.J. Hoff, H. Levanon, K. Möbius, R. Nechushtai, J.R. Norris, A. Scherz, J.L. Sessler, and D. Stehlik.
Isr. J. Chem. 32, 471-482 (1992).
|
|
|
Primary Events in Photosynthesis: Problems, Speculations, Controversies and Future Trends
M. Bixon, J. Fajer, G. Feher, J.H. Freed, D. Gamliel, A.J. Hoff, H. Levanon, K. Möbius, R. Nechushtai, J.R. Norris, A. Scherz, J.L. Sessler, and D. Stehlik.
Isr. J. Chem. 32, 471-482 (1992).
<doi: 10.1002/ijch.199200050>
Publication #209
|

|
|
INTRODUCTION: As was pointed out in Section 3.2.1, small g-factor differences for different magnetic "sites" or small g-factor anisotropies of the pigment radicals lead to strongly overlapping EPR lines at conventional X-band spectroscopy (9.5 GHz) for which even ENDOR∕TRIPLE does not result in well-resolved spectra. This is a common situation for photosynthetic preparations, in particular for the primary donor cation radical in RCs. In analogy to modern NMR spectroscopy, this problem can, in principle, be overcome by applying higher and higher magnetic fields. The increased Zeeman interaction ultimately leads, even for powder-type samples, to a complete separation of spectral features belonging to different principal values of the g-tensor. In addition to the improved Zeeman magnetoselection, high-field EPR is distinguished by its high detection sensitivity for small single crystals (high filling factor, see below). Another advantage of high-field EPR is the transformation of motionally narrowed spectra (ωeτR ≪ 1) into the slow-motion regime (ωeτR ≥ 1) with increased spectral sensitivity to motional dynamics (ωe is the Zeeman angular frequency and τR is the correlation time of the motion). High-field ENDOR has the additional advantage of allowing single-crystal-like spectra to be taken even from disordered solid state samples with small g-factor anisotropy, and of separating those ENDOR spectra from nuclei with different magnetic moments which overlap at X-band frequency.
Thanks to modern developments in ultra-high-frequency microwave technology, high-frequency∕high-field EPR spectrometers were built in various laboratories working, for instance, at 250 GHz (λ ≈ 1 mm, Bo ≈ 9 T for g = 2), at about 150 GHz (λ ≈ 2 mm), at 95 GHz (λ ≈ 3 mm), or at 70 GHz (λ ≈ 4 mm). So far the extension to ENDOR has been established up to the 3-mm band, but an ENDOR setup appears feasible also in the 1-mm band.
|
|
|
Spin Diffusion in Doubly Spin Polarized Atomic Deuterium
K.A. Earle, J.H. Freed and D.M. Lee.
J. Low Temp. Phys. 89, 911-937, (1992).
<doi: 10.1007/BF00683894>
Publication #208
|
|
|
ABSTRACT: Using an extension of the Boltzmann equation for the Wigner distribution appropriate for dilute spin 1 systems, spin diffusion equations are derived in the limit of large nuclear polarization in the non-degenerate régime. As an example of a system to which this work may be applied, the domain of validity of the Boltzmann equation for doubly spin-polarized deuterium, D↓↓, is studied. The effect of a finite field gradient is discussed. A calculated spin wave spectrum for a model one-dimensional system in the presence of a gradient is presented. Analogous effects in spin 1∕2 systems are compared and contrasted.
|
|
|
Critical Fluctuations and Molecular Dynamics at Liquid-Crystalline Phase Transitions. II. Electron Spin Resonance Experiments
A. Nayeem, S.B. Rananavare, V.S.S. Sastry, and J.H. Freed.
J. Chem. Phys. 96, 3912-3938 (1992).
<doi: 10.1063/1.461895>
Publication #207
|
|
|
ABSTRACT: Electron spin resonance (ESR) relaxation studies at nematic–isotropic (N–I), and nematic–smectic-A (N–SA) phase transitions in two liquid crystals, 4O,6 and 6OCB–8OCB, using the three spin probes, PD-tempone, MOTA, and P are described. In general, one finds that (i) at the N–I transition, as TNI is approached, the linewidths diverge with a critical exponent of 1∕2; (ii) at the N–SA transition, the linewidths diverge with a 1∕3 power law as the transition is approached from the nematic side. The nature of the critical divergences in the relaxation parameters is interpreted and analyzed in terms of fluctuations in the nematic and smectic order parameters at the respective transitions and the coupling of the orientational dynamics of the probe to these modes. Good quantitative agreement with theory for the N–I transition required the inclusion of the effects of asymmetric probe ordering. The theory developed in detail in paper I is applied to interpret the results at the N–SA transition. This theory is extended to include the effects of the measured anisotropies in (a) translational diffusion of the probe, (b) smectic correlation lengths, and (c) dynamic scaling exponents. In general, the magnitudes of the observed effects as well as their critical exponents are of the order expected, provided the averaging of the effects of density fluctuations within a smectic layer by probe diffusion is incomplete as a result of hindered diffusion.
|
|
|
Critical Fluctuations and Molecular Dynamics at Liquid-Crystalline Phase Transitions. I. Theoretical Aspects of the Nematic–Smectic-A Transition
J.H. Freed.
J. Chem. Phys. 96, 3901-3911 (1992).
<doi: 10.1063/1.461894>
Publication #206
|
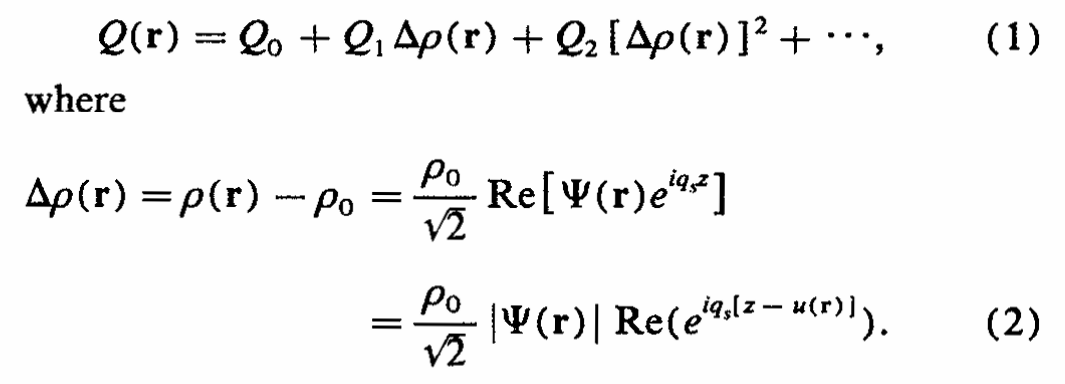
|
|
ABSTRACT: A theoretical model is developed for treating molecular dynamics at the nematic–smectic-A (N–SA) phase transition, which is frequently second order. This model is motivated by electron-spin-resonance (ESR) spin-relaxation studies of molecular probes. The critical dynamics of the hydrodynamic modes is described in accordance with dynamic scaling arguments of Brochard. Following Zager and Freed, the molecular dynamics of a probe molecule (governed by the molecular orientation and/or rotational diffusion) is assumed to couple to fluctuations in the smectic order parameter, because these molecular properties are a function of the precise location of the probe within the transient smecticlike layer. Two limiting cases of (1) (nearly) free translational diffusion of the probe across the smecticlike layer; and (2) expulsion of the probe to the aliphatic chains with highly hindered diffusion (i.e., jump diffusion) across the smecticlike layer are considered. The relevant spectral density shows critical types of divergence, where the exponent depends strongly on the details of the model. It is found that only the (near) zero-frequency spectral densities can show such divergences. It is pointed out that spectral densities available for spin relaxation do not truly diverge as the N–SA transition is approached arbitrarily closely, because ultimately motional-narrowing theory will no longer be valid, and fluctuations begin to be frozen on the ESR time scale. This matter is briefly analyzed. Also considered briefly are the effects of anisotropies in the smectic phase and of fluctuations in nematic director near the N–SA transition.
|
|
|
A Many-Body Stochastic Approach to Rotational Motions in Liquids
A. Polimeno and J.H. Freed.
Adv. Chem. Phys. 83, 89-206 (1993).
<doi: 10.1002/9780470141410.ch3>
Publication #205
|
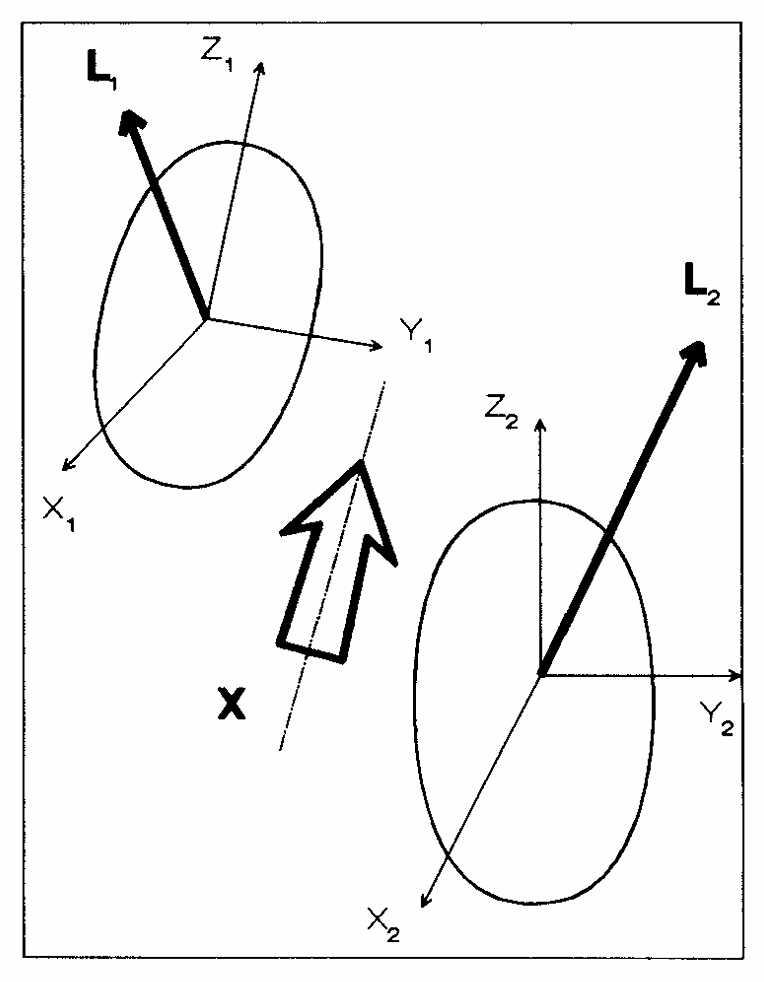
|
|
INTRODUCTION: Classic Brownian motion has been widely applied in the past to the interpretation of experiments sensitive to rotational dynamics. ESR and NMR measurements of T1 and T2 for small paramagnetic probes have been interpreted on the basis of a simple Debye model, in which the rotating solute is considered a rigid Brownian rotator, such that the time scale of the rotational motion is much slower than that of the angular momentum relaxation and of any other degree of freedom in the liquid system. It is usually accepted that a fairly accurate description of the molecular dynamics is given by a Smoluchowski equation (or the equivalent Langevin equation), that can be solved analytically in the absence of external mean potentials.
Since the pioneering contribution of Debye, one-body Smoluchowski equations have provided a general framework for the study of dielectric relaxation in liquids, neutron scattering, and infrared spectroscopy. The basic hypothesis is that the solute degrees of freedom are the only "relevant" (i.e., slow when compared with the timescale of the experiment) variables in the system, and that the surrounding liquid medium behaves as a homogeneous bath whose internal degrees of freedom are rapidly relaxing. This simple picture has had many substantial refinements and improvements. Perrin, Sack, Fixman and Rider, Hubbard, McClung, Morita, and many others have contributed by including anisotropy and inertial effects and by studying detailed numerical solutions to classic Fokker–Planck–Kramers equations for the tumbling of a general top. Good agreement between the experimental data and theoretical predictions can often be obtained at moderate viscosities and pressures. Also, the influence of a mean potential of interaction has been extensively studied, since the original work of Favro.
|
|
|
Spatially Resolved Two-Dimensional Fourier Transform Electron Spin Resonance
U. Ewert, R.H. Crepeau, S. Lee, C.R. Dunnam, D. Xu, and J.H. Freed.
Chem. Phys. Lett. 184, 34-40 (1991).
<doi: 10.1016/0009-2614(91)87160-D>
Publication #204
|
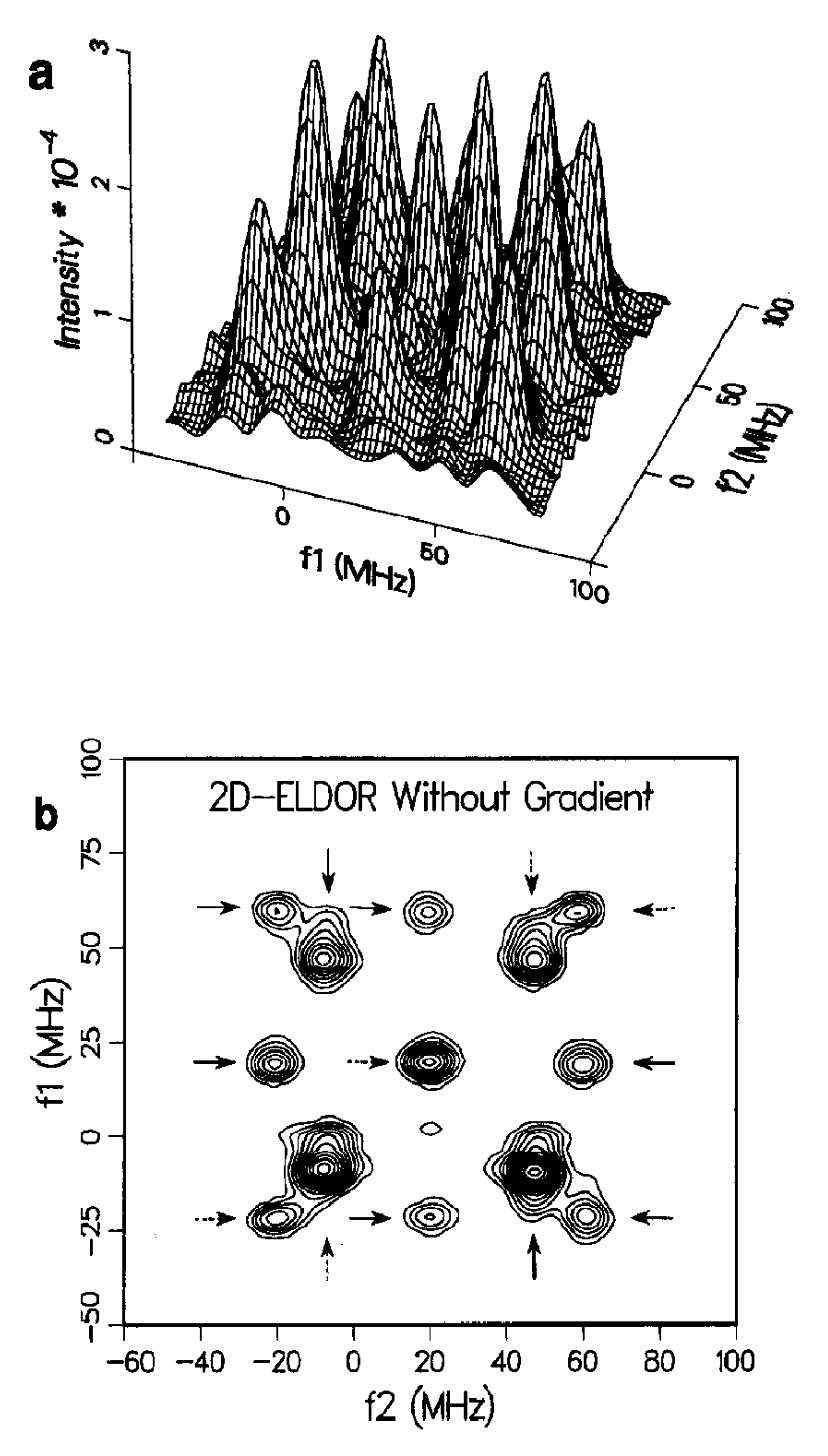
|
|
ABSTRACT: Fourier transform ESR methods have been extended to permit spatially resolved two-dimensional (2D)-ESR experiments. This is illustrated for the case of 2D-electron-electron double resonance (2D-ELDOR) spectra of nitroxides in a liquid that exhibits appreciable cross-peaks due to Heisenberg spin exchange. The use of spin-echo decays in spatially resolved FT-ESR is also demonstrated.
|
|
|
Fourier Transform Electron Spin Resonance Imaging
U. Ewert, R.H. Crepeau, C.R. Dunnam, D. Xu, S. Lee, and J.H. Freed.
Chem. Phys. Lett. 184, 25-33 (1991).
<doi: 10.1016/0009-2614(91)87159-9>
Publication #203
|
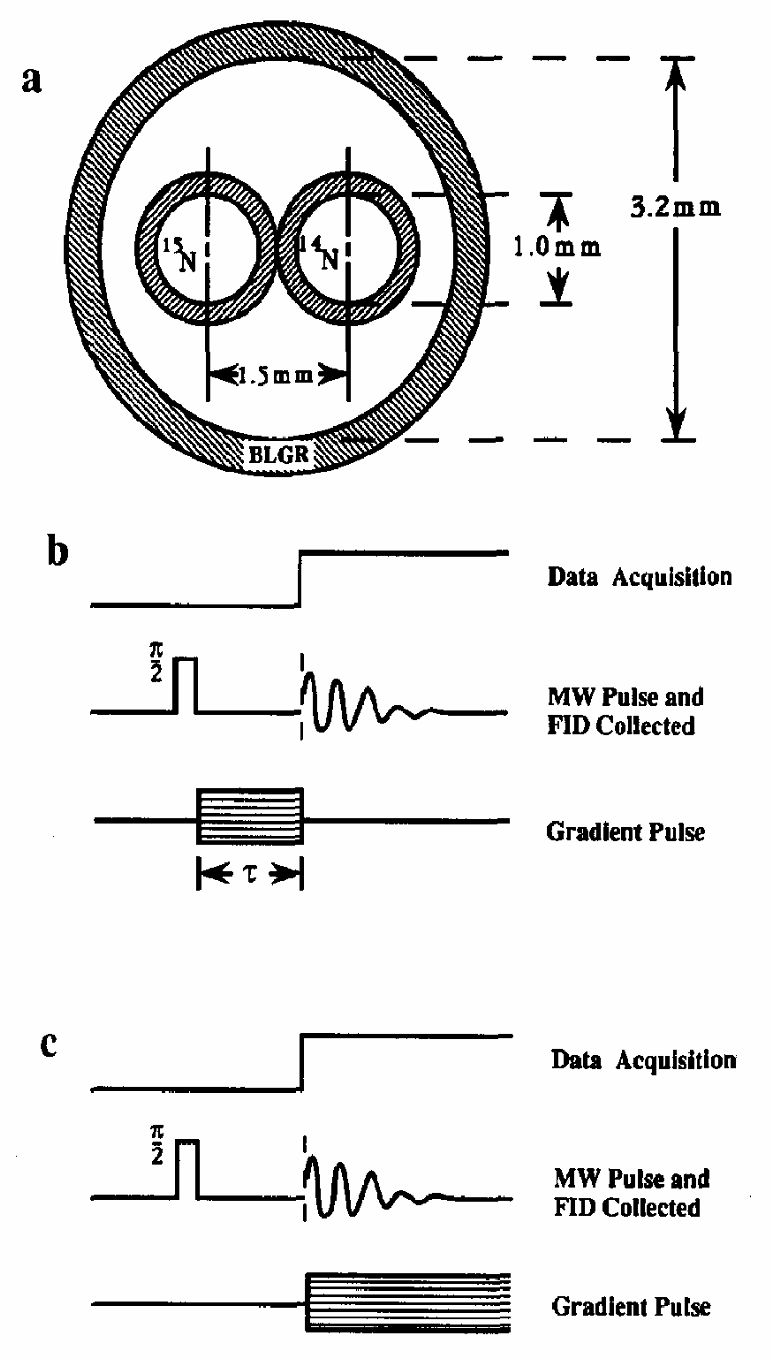
|
|
ABSTRACT: Modern Fourier transform (FT) ESR methods have been combined with fast, high power pulsed magnetic field gradients to enable FT-ESR imaging. Spectral—spatial imaging by frequency and phase encoded FT methods are compared with cw methods. The initial phase encoded results are comparable in quality to those from the well-developed cw methods and further improvements which would enhance FT-ESR imaging are noted.
|
|
|
Two-Dimensional Fourier Transform ESR in the Slow-Motional and Rigid Limits: 2D-ELDOR
B.R. Patyal, R.H. Crepeau, D. Gamliel, and J.H. Freed.
Chem. Phys. Lett. 175, 453-460 (1990).
<doi: 10.1016/0009-2614(90)85563-R>
Publication #202
|
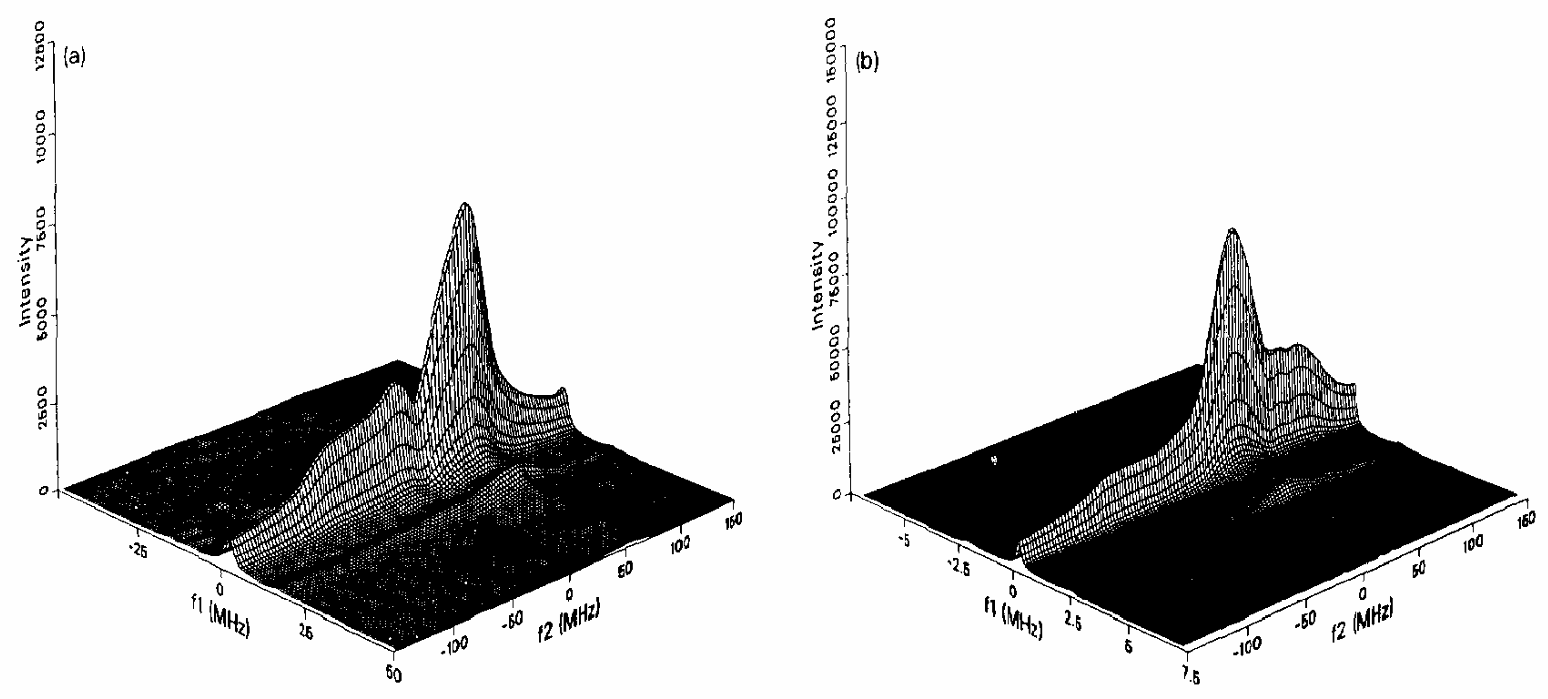
|
|
ABSTRACT: The two-dimensional Fourier transform ESP techniques of stimulated SECSY and 2D-ELDOR are shown to be powerful methods for the study of slow motions for nitroxides. Stimulated SECSY provides magnetization transfer rates, whereas 2D-ELDOR displays how the rotational motions spread the spins out from their initial spectral positions to new spectral positions, as a function of mixing time. The role of nuclear modulation in studies of structure and dynamics is also considered.
|
|
|
Two-Dimensional Fourier Transform ESR in the Slow-Motional and Rigid Limits: SECSY-ESR
B.R. Patyal, R.H. Crepeau, D. Gamliel, and J.H. Freed.
Chem. Phys. Lett. 175, 445-452 (1990).
<doi: 10.1016/0009-2614(90)85562-Q>
Publication #201
|
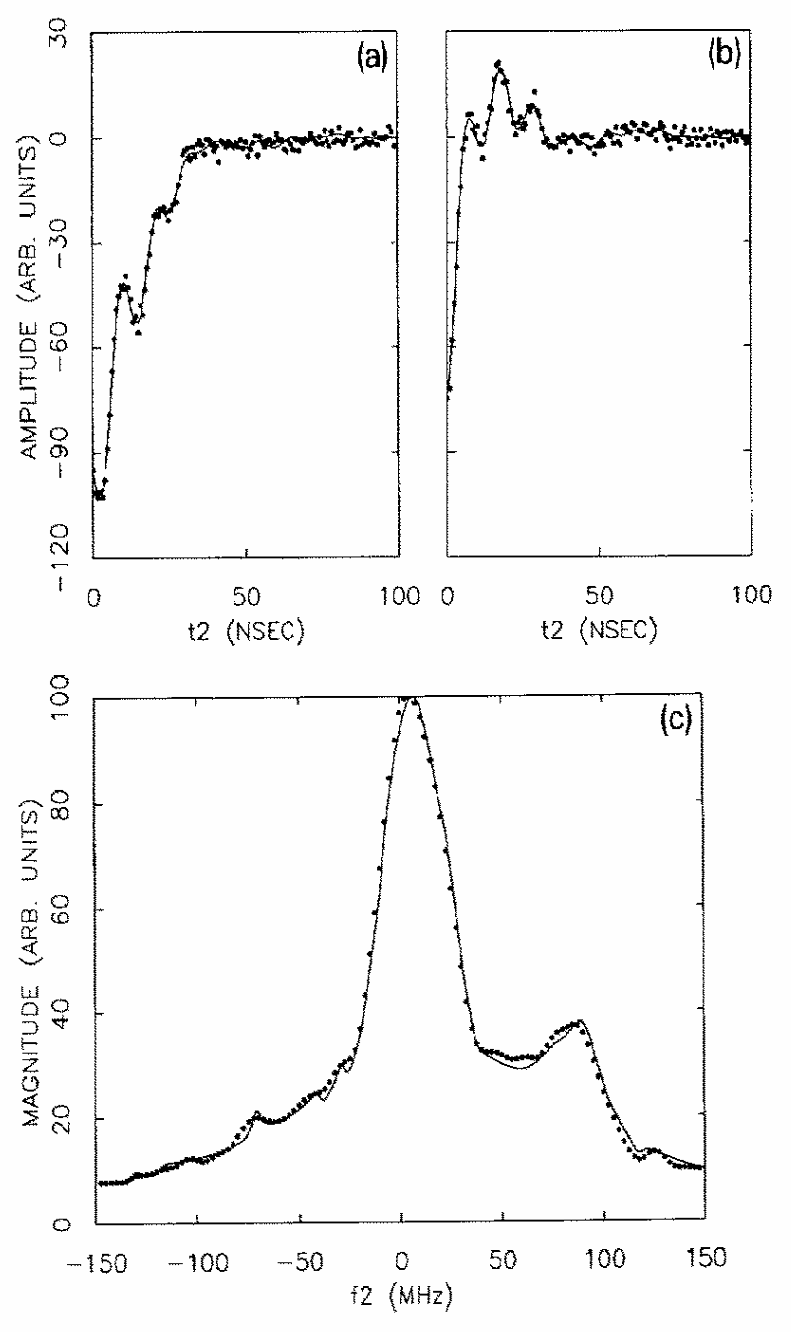
|
|
ABSTRACT: Two-dimensional Fourier transform ESR has now been extended to the slow-motional and rigid limit regimes which require spectral bandwidths of 200–250 MHz and sub-nanosecond time resolution in echo decays. Two-pulse SECSY-ESR on nitroxides is shown to provide detailed structural information from the effects of nuclear modulation and slow-motional information from both the main ESR spectrum as well as the nuclear modulation.
|
|
|
Microscopic Versus Macroscopic Diffusion in Model Membranes by Electron Spin Resonance Spectral-Spatial Imaging
Y.-K. Shin, U. Ewert, D.E. Budil, and J.H. Freed.
Biophys. J. 59, 950-957 (1991).
<doi: 10.1016/S0006-3495(91)82310-4>
PMID:
1648417
PMCID:
PMC1281263
Publication #200
|
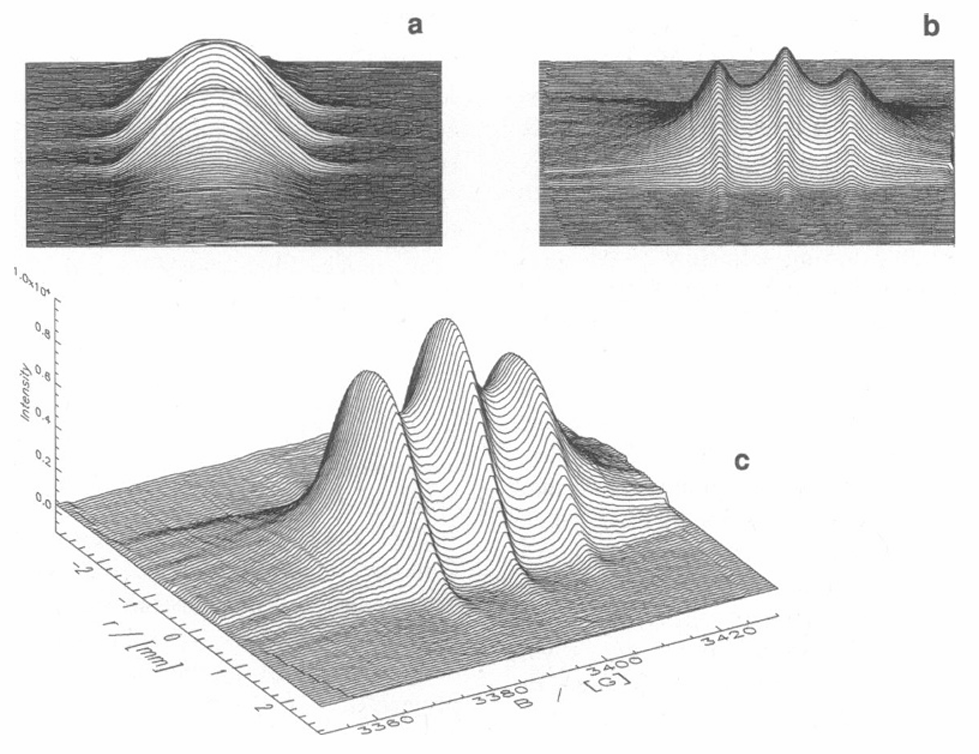
|
|
ABSTRACT: The macroscopic and the microscopic diffusion coefficients of a phospholipid spin label (16-PC) in the model membrane 1-palmitoyl-2-oleoyl-sn-glycero-phosphatidylcholine have been measured simultaneously in the same sample utilizing the new technique of spectral-spatial electron spin resonance imaging. The macroscopic diffusion coefficient Dmacro for self-diffusion of 16-PC spin label is obtained from imaging the concentration profiles as a function of time, and it is (2.3 ± 0.4) × 10-8 cm2∕s at 22°C. The microscopic diffusion coefficient Dmicro for relative diffusion of the spin probes is obtained from the variation of the spectral line broadening with spin label concentration, which is due to spin-spin interactions. Dmicro is found to be substantially greater than Dmacro for the same sample at the same conditions, and is estimated to be at least (1.0 ± 0.4) × 10-7 cm2∕s. Possible sources for their difference are briefly discussed in terms of the models used for Dmicro.
|
|
|
|
|
|
|
Non-Resonant Microwave Absorption in High-Tc Thin Films
R. Durny, A. Dulčić, R.H. Crepeau, J.H. Freed and P. Kus.
Physica C 171, 401-405 (1990).
<doi: 10.1016/0921-4534(90)90248-D>
Publication #197
|
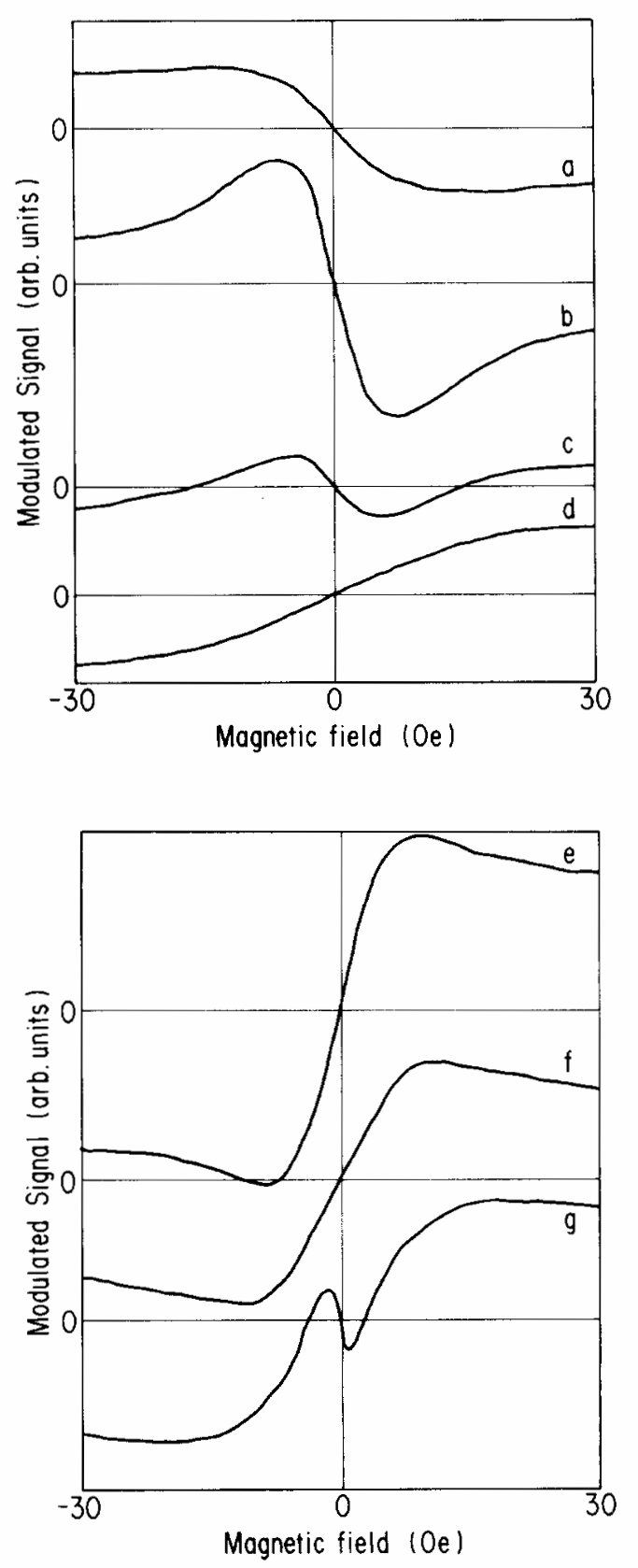
|
|
ABSTRACT: Magnetic-field-dependent non-resonant microwave absorption in thin film samples of various high-Tc superconductors is reported. Complex types of signals were observed as the temperature was lowered from Tc to ˜ 10 K. Possible correlation between the thin film quality and the occurrence of the signals is suggested.
|
|
|
The Bruker Lecture: Modern Techniques in Electron Paramagnetic Spectroscopy
J.H. Freed.
J. Chem. Soc. Faraday Trans. 86, 3173-3180 (1990).
<doi: 10.1039/FT9908603173>
Publication #196
|
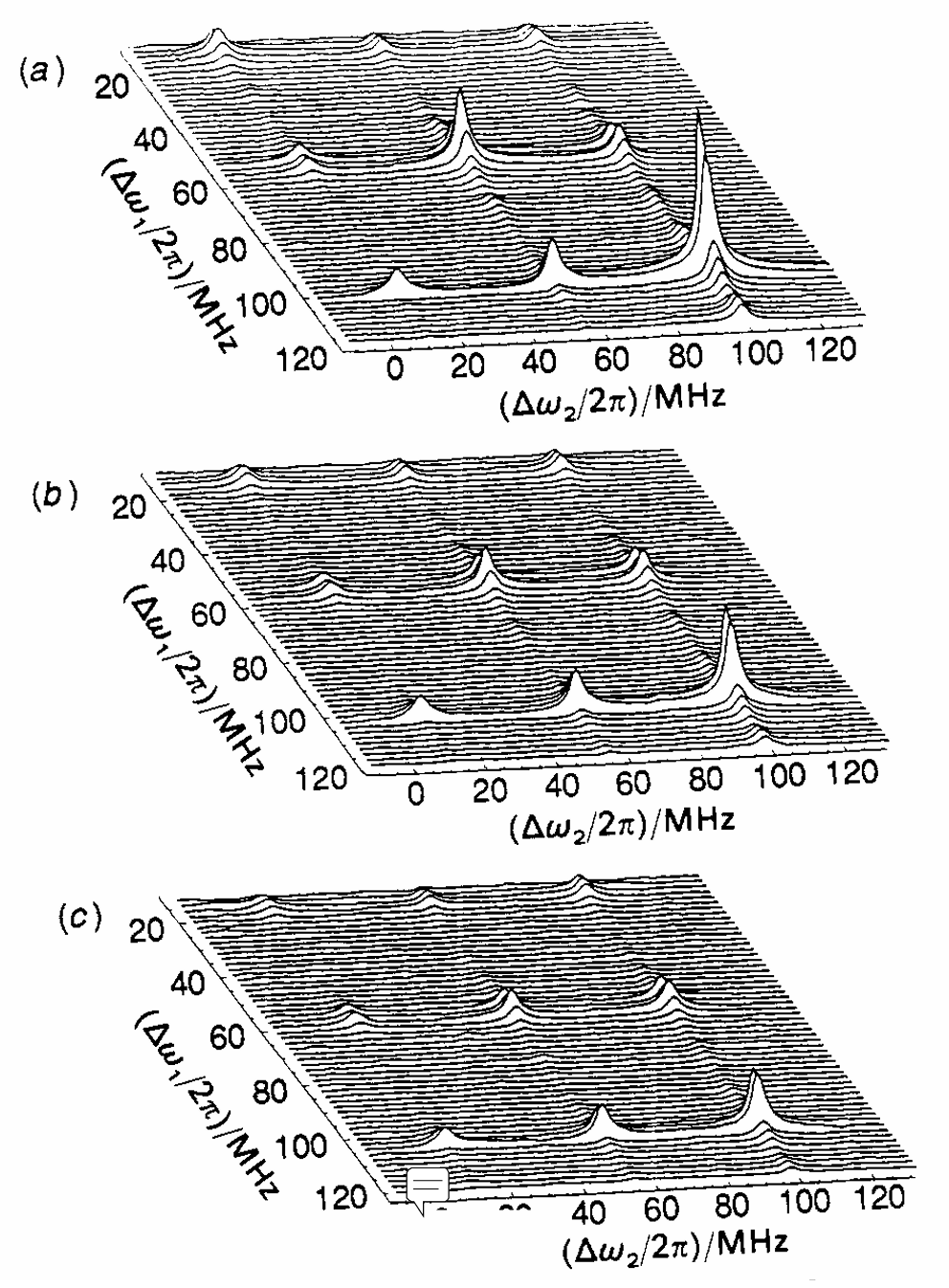
|
|
ABSTRACT: The last few years have seen important new advances in EPR which have the possibility of revolutionizing the applications of EPR in chemistry and related fields. New methods include (a) two-dimensional and Fourier-transform EPR; (b) far-infrared EPR (with 1 mm waves); (c) dynamic imaging of diffusion by EPR; (d) powerful computational algorithms for spectral simulation. These new methods and their implications for chemical research are discussed.
|
|
|
ESR Studies of Stearic Acid Binding to Bovine Serum Albumin
M. Ge, S.B. Rananavare, and J.H. Freed.
Biochim. Biophys. Acta 1036, 228-236 (1990).
<doi: 10.1016/0304-4165(90)90039-Y>
Publication #195
|

|
|
ABSTRACT: The ESR spectra of a series of chain-labelled doxyl stearic acids (5-, 7-, 12- and 16-DSA) and doxyl methyl stearates (5-, 7-, 12- and 16-DMS) bound to the high-affinity binding sites of bovine serum albumin (BSA) have been analyzed using nonlinear least-squares fitting of slow-motional ESR simulation. The motional analysis reveals that the rotational diffusion of these stearates around the axis perpendicular to the long hydrocarbon chain is greatly hindered, suggesting that they are held tightly in a channel of the protein. Comparison of the isotropic hyperfine splitting, A0, among each series shows that 5- and 16-DSA and 16-DMS have larger A0 values than the other spin labels. In addition, labels at the 16-C position of both DSA and DMS exhibit significantly increased motion relative to the other positions. These observations suggest that the channel starts at 5-C of the chain and ends somewhere between 13-C and 15-C, leading to an estimate of 11 ± 1Å for the lenght of the channel. The methyl stearate lables exhibit significantly faster rotation around the chain axis than the analogous stearic acid labels, suggesting a double hydrogen-bonding mechanism for fatty acid binding to BSA. The ability of the acid to form two hydrogen bonds apparently fixes it more rigidly in the protein preventing rotation about either single hydrogen bond. A double-hydrogen bonding mechanism is most consistent with the formation of a salt bridge between the negatively charged carboxylate of the acid and either a positively charged guanidino group of arginine, or the positively charged ω-amino groups of two lysine residues. An ESR study of the pH dependence of DSA binding indicates that salt bridge formation with lysine is responsible for at least some of the long chain fatty acid binding sites of BSA.
|
|
|
Weak-link structure in YBa2Cu3O7 single crystals: A microwave study
A. Dulčić, R. H. Crepeau, J. H. Freed, L. F. Schneemeyer and J. V. Waszczak
Phys. Rev. B 42, 2155-2160 (1990).
<doi: 10.1103/PhysRevB.42.2155>
Publication #194
|
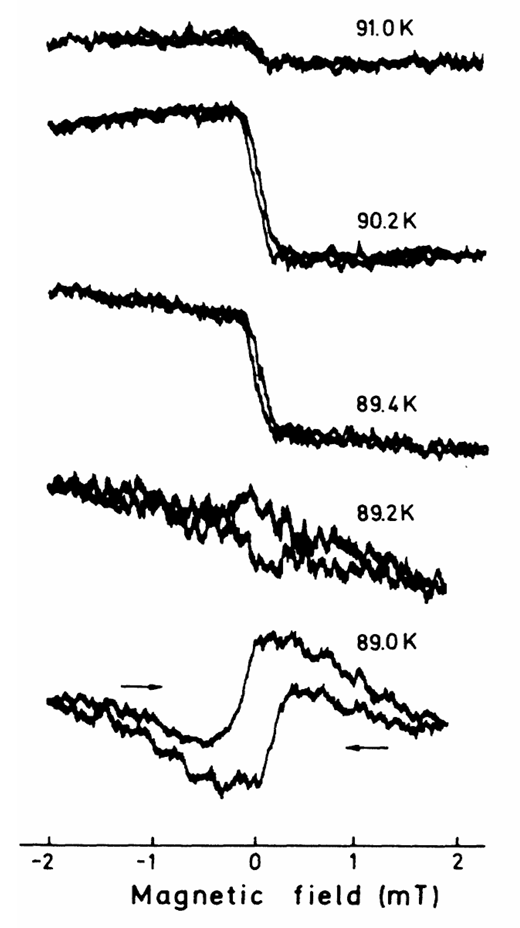
|
|
ABSTRACT: Field-dependent microwave absorption in high-quality YBa2Cu3O7 single crystals shows that even very low magnetic fields penetrate into the sample. In the temperature interval of a few degrees below Tc, a broad hysteretic signal is observed. We interpret it in terms of a dense structure of weak links, whose origin is in twin boundaries and other thin defect planes. As the temperature is reduced, the critical currents of these junctions increase rapidly, and approach the bulk value. The broad signal then vanishes. At low temperatures one can observe narrow discrete absorption lines due to occasional defects of a different kind, which are thicker and, therefore, maintain a weak coupling. The dense structure of weak links, active just below Tc, may preclude the measurements of some intrinsic bulk superconducting properties close to Tc, and invalidate a straightforward comparison of experimental data with theoretical predictions.
|
|
|
|
|
ESR Studies of Molecular Dynamics at Phase Transitions in Liquid Crystals
J.H. Freed, A. Nayeem, and S.B. Rananavare.
In The Molecular Dynamics of Liquid Crystals; G. Luckhurst and C. Veracini, Eds.
Kluwer: Dordrecht, Netherlands, 1994; Chapter 14, pp. 335-364.
<doi: 10.1007/978-94-011-1168-3_14>
Publication #192
|
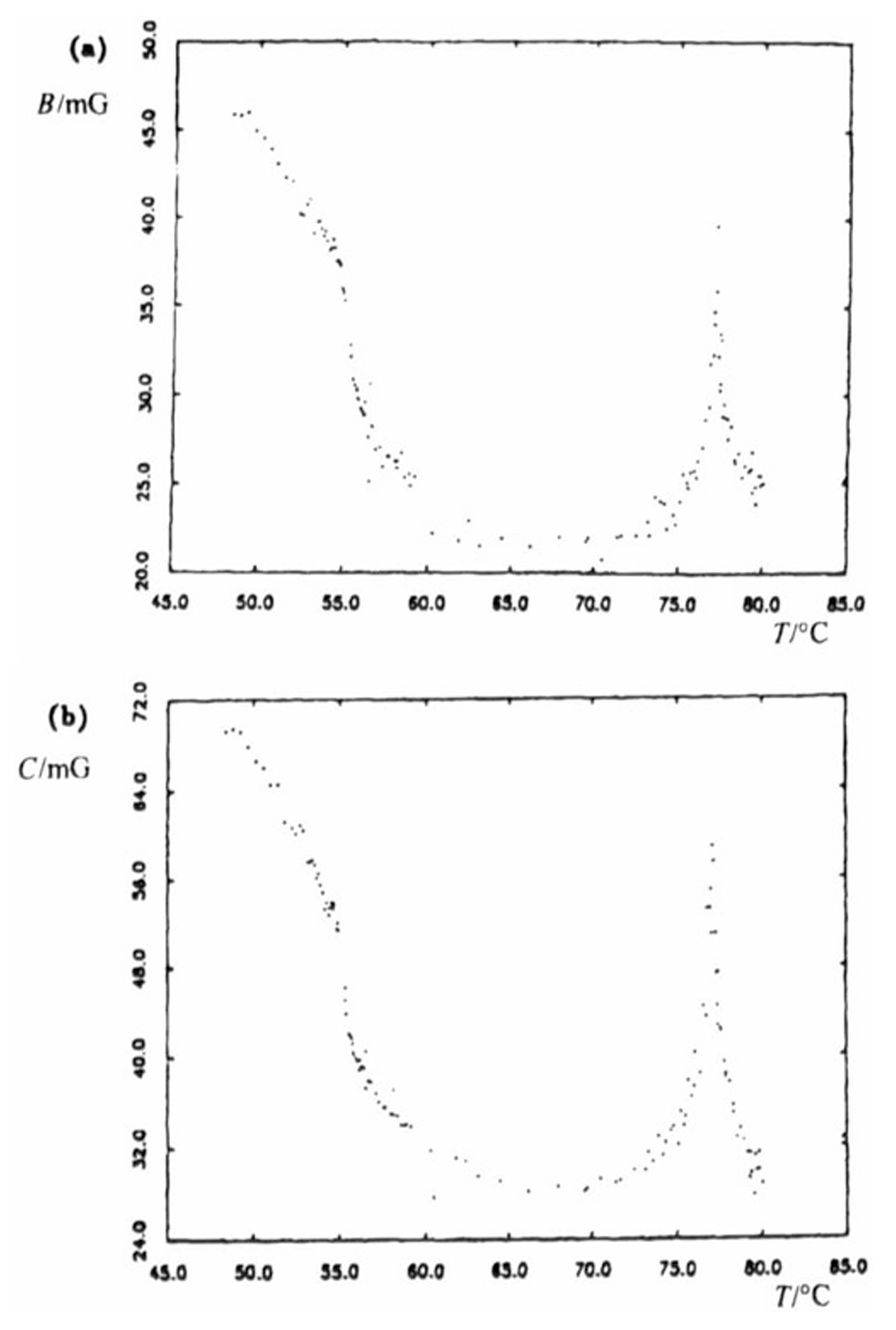
|
|
ABSTRACT: Order parameter fluctuations at mesomorphic phase transitions modulate the molecular dynamics of spin probes, thereby affecting spin relaxation. The anomalous relaxation rates at the phase transitions (T*) often diverge as ∣T - T*∣γ, where γ is a critical exponent, and has been noted to be universal. For N-I transitions, γ = -½, and for SA−N transitions, γ = −⅓. Theoretical models are discussed that provide a unified framework for rationalising the experimental results.
|
|
|
ESR and Slow Motions in Liquid Crystals
J.H. Freed, A. Nayeem, and S.B. Rananavare.
In The Molecular Dynamics of Liquid Crystals; G. Luckhurst and C. Veracini, Eds.
Kluwer: Dordrecht, Netherlands, 1994; Chapter 15, pp. 365-402.
|
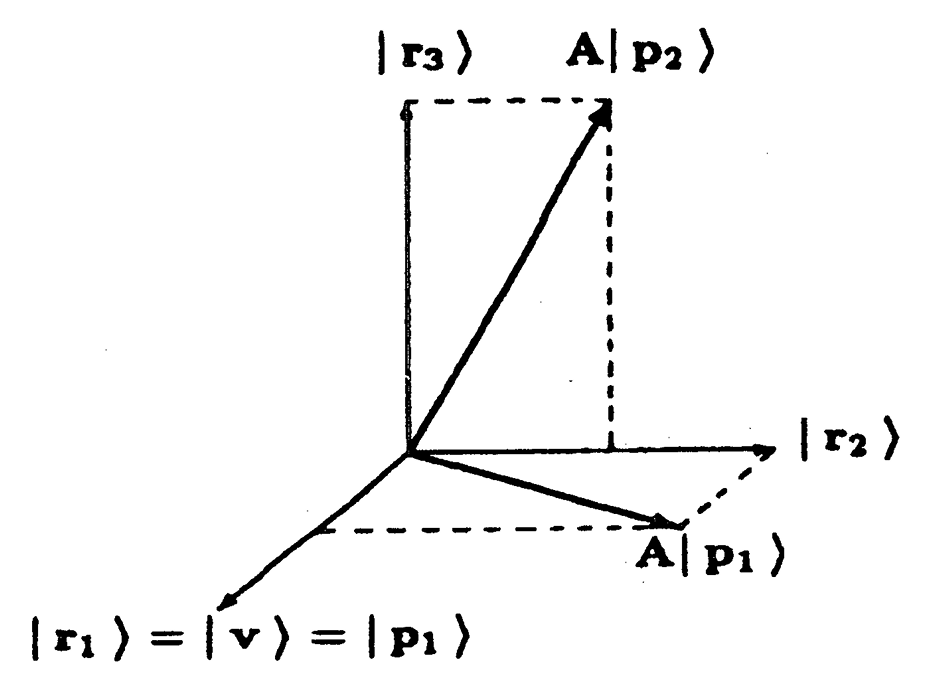
|
|
ESR and Slow Motions in Liquid Crystals
J.H. Freed, A. Nayeem, and S.B. Rananavare.
In The Molecular Dynamics of Liquid Crystals; G. Luckhurst and C. Veracini, Eds.
Kluwer: Dordrecht, Netherlands, 1994; Chapter 15, pp. 365-402.
<doi: 10.1007/978-94-011-1168-3_15>
Publication #191
|

|
|
ABSTRACT: The theory of slow motional ESR is outlined, with applications to nitroxide spectra in liquid crystals. The computer algorithms for implementing the theory based on the stochastic Liouville equation are also discussed.
|
|
|
ESR and Molecular Motions in Liquid Crystals: Motional Narrowing
J.H. Freed, A. Nayeem, and S.B. Rananavare.
In The Molecular Dynamics of Liquid Crystals; G. Luckhurst and C. Veracini, Eds.
Kluwer: Dordrecht, Netherlands, 1994; Chapter 12, pp. 271-312.
<doi: 10.1007/978-94-011-1168-3_12>
Publication #190
|
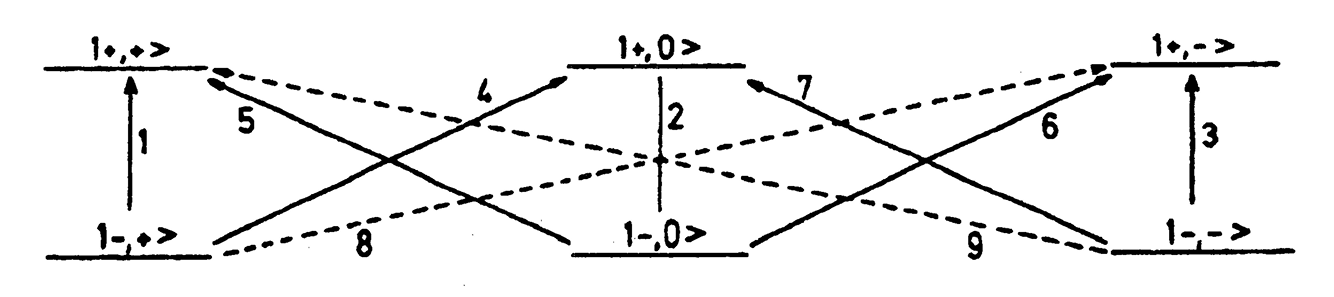
|
|
ABSTRACT: Theoretical and experimental aspects of spin relaxation in liquid crystals are considered here, with primary emphasis on the motional narrowing regime. ESR studies of translational motion in mesophases are also described.
|
|
|
|
|
Theory of Two-Dimensional ESR with Nuclear Modulation
D. Gamliel and J.H. Freed.
J. Magn. Reson. 89, 60-93 (1990).
<doi: 10.1016/0022-2364(90)90162-3>
Publication #188
|
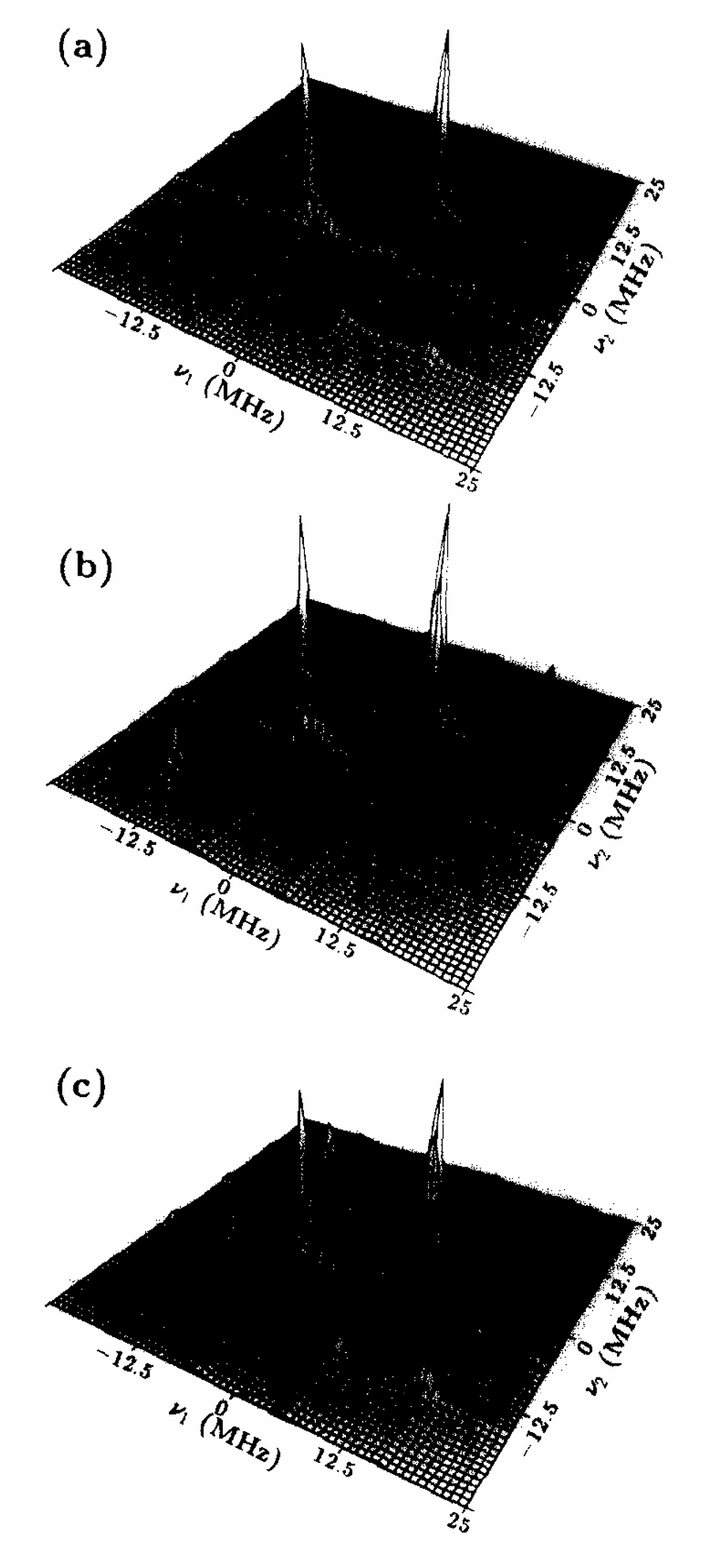
|
|
ABSTRACT: A formalism for computing 2D ESR lineshapes with nuclear modulation is developed in a form which is useful for planning phase cycles for particular purposes. A simple method of processing spectra, utilizing quadrature detection, is shown to enhance the selectivity of the phase cycling techniques. Computed ESR-COSY, ESR-SECSY, and 2D ELDOR lineshapes are presented for several kinds of polycrystalline and single-crystal samples which exhibit nuclear modulation, due to one or several nuclei. The two-dimensional methods are found to give more detailed structural information than the corresponding ESEEM spectra. New phase cycles are found to eliminate completely all transverse and axial peaks in 2D ELDOR and in ESR-COSY, and at the same time eliminate all artifacts arising from incomplete image rejection. Other phase cycles are presented for selecting in those experiments only axial peaks, for measuring T1. It is also shown how selective phase cycles may help to distinguish between coherent and exchange cross peaks. In the special case of nitroxides in typical Zeeman fields, there are no significant nuclear modulation effects from the 14N nuclear spin interaction, but those from the protons (or deuterons) will, in general, be significant.
|
|
|
Dynamics of Phosphatidylcholine-Cholesterol Mixed Model Membranes in the Liquid Crystalline State
Y.K. Shin, J.K. Moscicki, and J.H. Freed.
Biophys. J. 57, 445-459 (1990).
<doi: 10.1016/S0006-3495(90)82561-3>
PMID:
2155032
PMCID:
PMC1280739
Publication #187
|
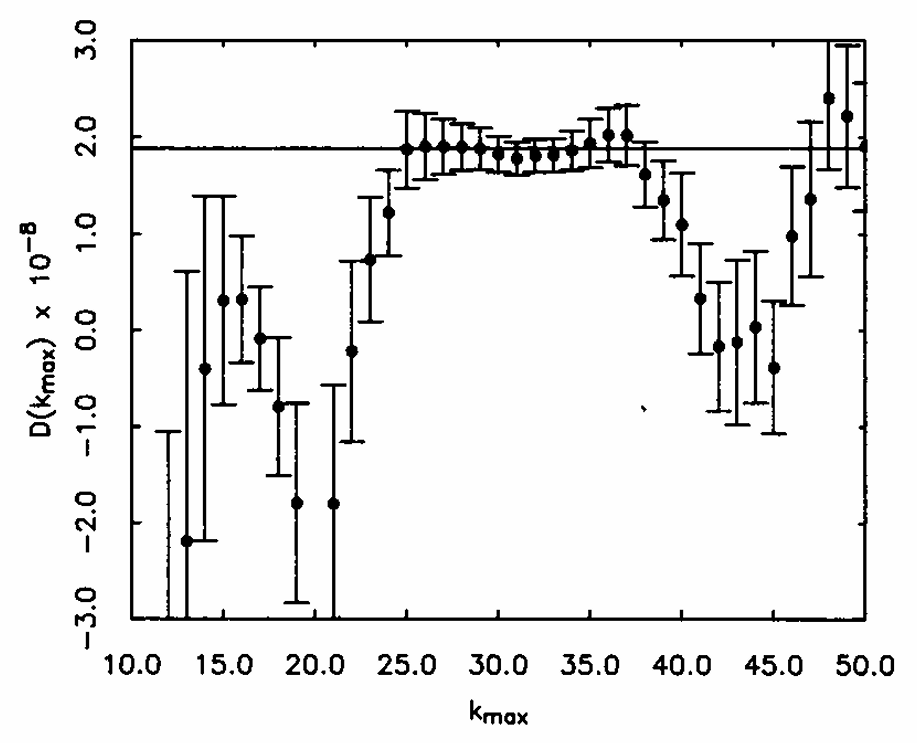
|
|
ABSTRACT: The effects of cholesterol on the dynamics of cholestane spin probe (CSL) in various phosphatidylcholine-cholesterol mixed model membranes are examined. The lateral diffusion, D of CSL in DMPC∕POPC∕cholesterol ternary mixtures, is measured utilizing an improved dynamic imaging electron spin resonance method. It shows a factor of two decrease at 10 mol % and 25°C, whereas it shows only a 40% decrease at 50 mol % and 50°C. A comparison with results in POPC∕cholesterol mixtures, which show a stronger effect of cholesterol on D, indicates that acyl chain unsaturation leads to stronger self association of cholesterol in PC model membranes. An S2CSL dependence of the activation energy for D, has been confirmed for the DMPC∕POPC∕cholesterol mixtures. Here SCSL is the order parameter for CSL. A similar correlation of R⊥, the perpendicular component of the rotational diffusion coefficient, with SCSL, which is true for all three mixtures (DMPC∕cholesterol, POPC∕cholesterol, and DMPC∕POPC∕cholesterol) we have studied, is also found. These are associated with the effects of enhanced local ordering on the free volume needed for translation and reorientation. Such correlations of dynamic properties D and R⊥ with the thermodynamic quantity S, as well as the consistent interpretations of the effect of acyl chain unsaturation on the dynamics in terms of the activity coefficients, strongly emphasize the interrelation between the dynamic structure and the thermodynamics of the PC∕cholesterol mixtures.
|
|
|
The Method of Dynamic Imaging of Diffusion by EPR
J.K. Moscicki, Y.K. Shin, and J.H. Freed.
In EPR Imaging and In Vivo EPR; G.R. Eaton, S.S. Eaton, and K. Ohno, Eds.
CRC Press: New York, NY, 1991; Chapter 19, pp. 189-219.
Publication #186
|
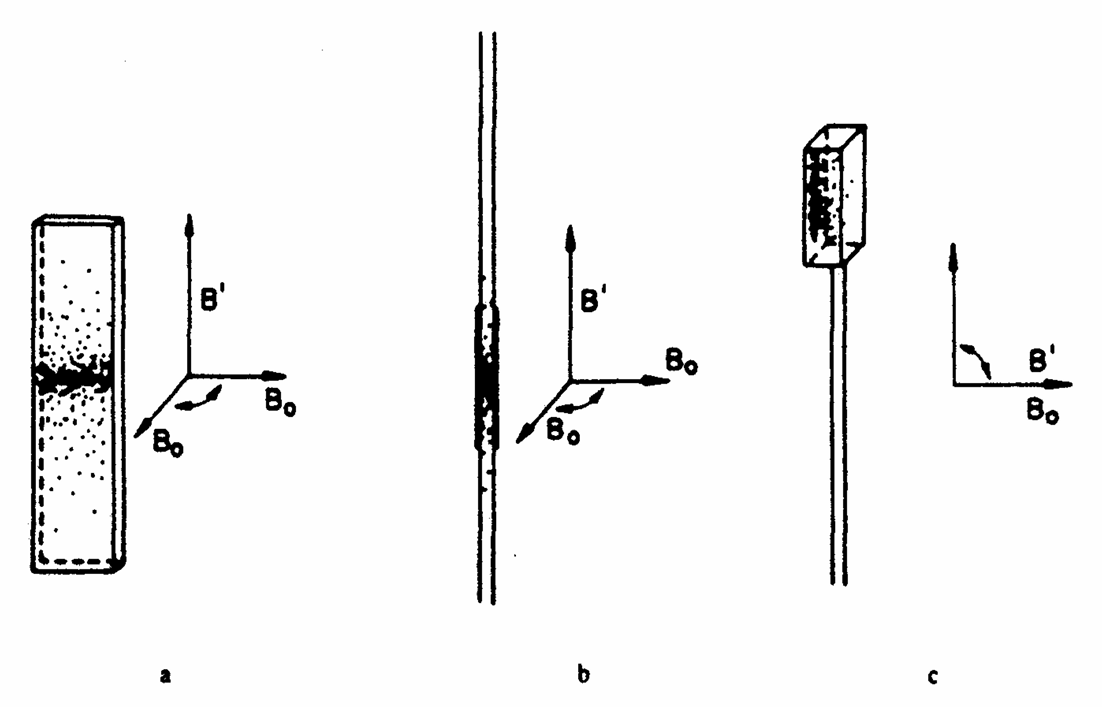
|
|
INTRODUCTION: Both thermotropic and lyotropic liquid crystals appeal to scientists for their unique properties of being more or less ordered while at the same time preserving a high degree of molecular mobility. Furthermore, a number of lyotropic liquid-crystalline phases show space structures relevant to biological systems. Not surprisingly, therefore, studies of the molecular dynamics in mesomorphic states of condensed matter have attracted a sustained interest over the past several years. The most fundamental characteristic of liquid-crystalline states, at least from a microscopic point of view, is the presence of long-range orientational order, while positional order is limited or absent altogether. A first step towards the understanding of the relation between molecular properties and the macroscopic structure of mesophases consists of collecting information about the local behavior of the molecule subject to a mean-ordering potential. Consequently, the rotational dynamics was vigorously studied in the past. Studies were facilitated by the existence of several techniques sensitive to molecular reorientations in external fields (dielectric relaxation, FIR, Raman, nuclear magnetic resonance or NMR, electron paramagnetic resonance or EPR, and other spectroscopies). Progress in translational diffusion measurements was much slower, despite its importance for understanding the anisotropy of mass transport, critical phenomena at liquid-crystalline phase transitions, and mass transport in model and biomembranes. It was essentially due to the lack of reliable experimental techniques enabling such studies in liquid-crystalline materials.
One can divide experiments designed to measure the translational diffusion constant, D, into two general categories. A "macroscopic" method involves diffusion over distances, several orders of magnitude larger than molecular dimensions, whereas a "microscopic" method measures diffusion over dimensions on the order of molecular lengths. Early efforts on mesomorphic materials were essentially restricted to the first category and involved impurity diffusion across the sample. These include chemical, optical, and radioactive probes, charge carriers, and NMR with pulsed gradients. In the last decade, there was a rapid development in experimental techniques which permit the diffusion coefficient to be measured spectroscopically. Macroscopic diffusion coefficients are directly measured from NMR field-gradient spin echoes. Macroscopic techniques employed to measure diffusion of spin probes and spin labels include that of Sheats and McConnell which requires selective photobleaching of a sample and that of Ahn which applies the capillary-diffusion method to EPR and requires a great deal of measurement time. Recently, EPR imaging has been intensively applied to study mass transport in liquids, thermotropic liquid crystals, model membranes, and biologically relevant polymers.
|
|
|
Dynamic Imaging of Diffusion in One Dimension by cw-ESR
J.K. Moscicki, Y.K. Shin, and J.H. Freed.
Physica Medica V, 229-237 (1989).
Publication #185
|
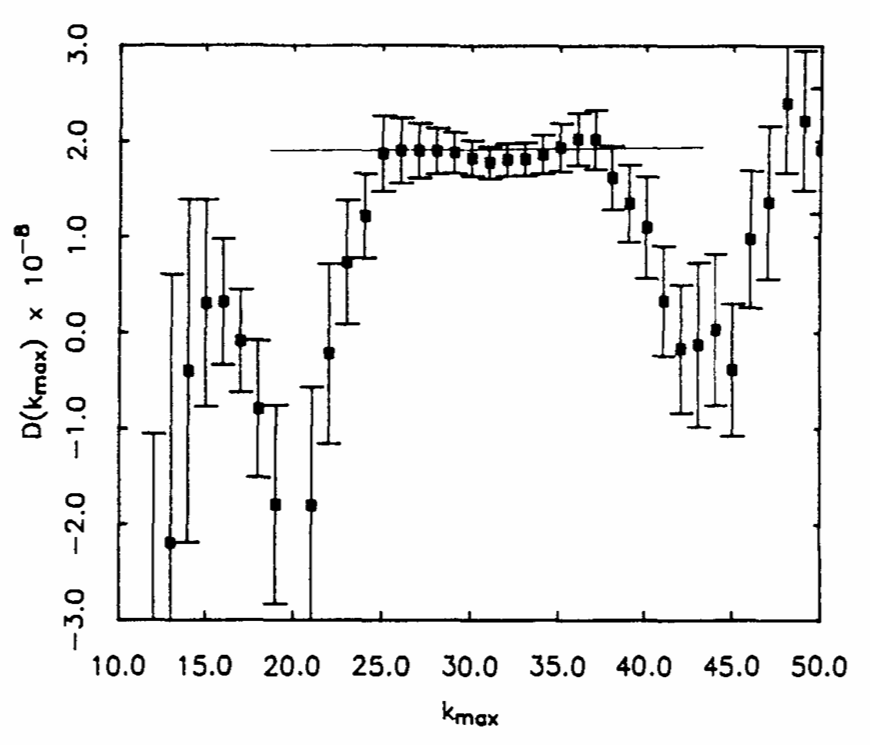
|
|
ABSTRACT: Dynamic Imaging of Diffusion by ESR has in the last several years emerged as a powerful new technique for accurate measurement of the translational diffusion coefficients of nitroxide radicals in isotropic and oriented (anisotropic) fluids. The methodology and some recent results on oriented model membranes are reviewed in this work.
|
|
|
Thermodynamics of Phosphatidylcholine-Cholesterol Mixed Model Membranes in the Liquid Crystalline State Studied by the Orientational Order Parameter
Y.K. Shin and J.H. Freed.
Biophys. J. 56, 1093-1100 (1989).
<doi: 10.1016/S0006-3495(89)82757-2>
PMID:
2558733
PMCID:
PMC1280613
Publication #184
|
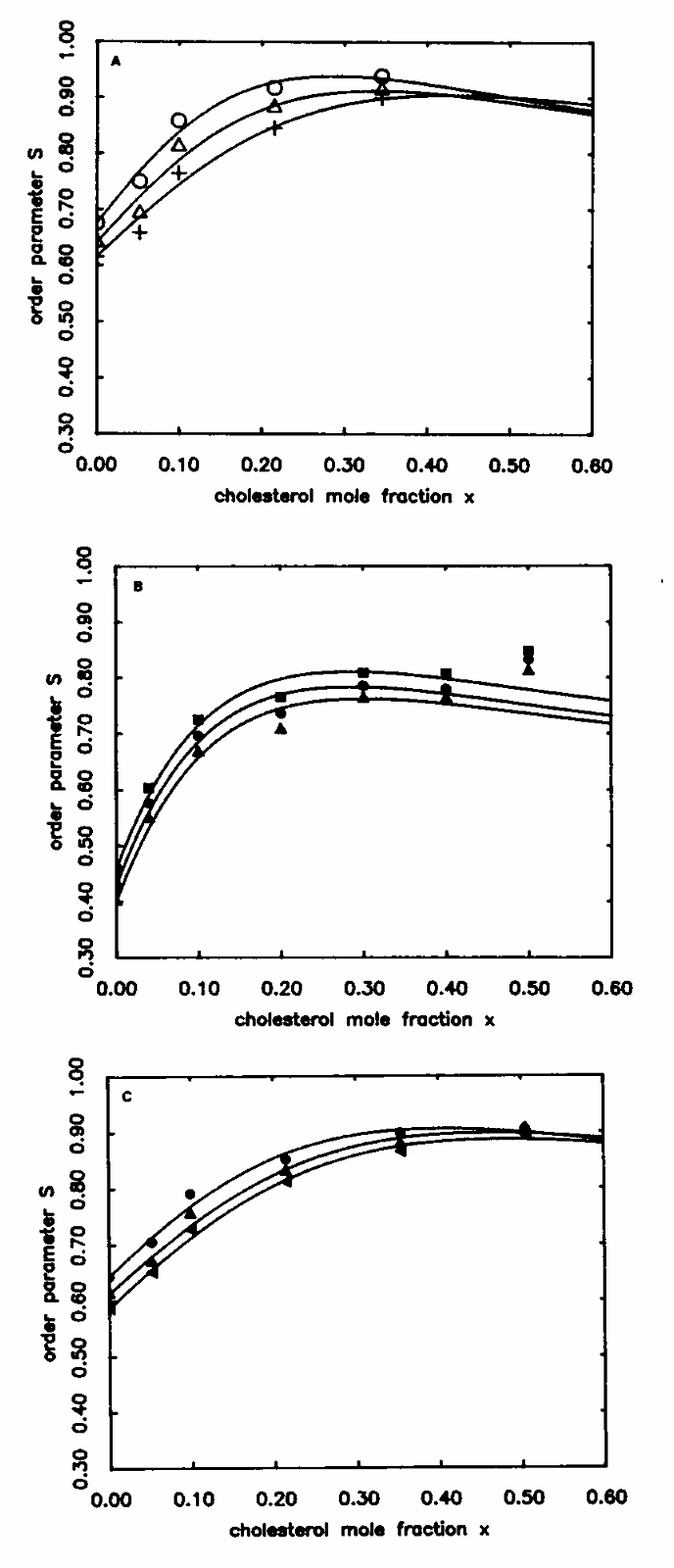
|
|
ABSTRACT: It is shown that good estimates of the activity of cholesterol in phosphatidylcholine-cholesterol mixed model membranes are obtained by examining the orientational order parameter S of cholestane spin probe (CSL) that is obtained from electron spin resonance by spectral simulation. By introducing thermodynamic stability conditions of liquid mixtures, the variation of activity (or S) as a function of cholesterol mole fraction is utilized to predict the concentration at which the phase separation occurs. These results for DMPC and cholesterol binary mixtures agree very well with those of Tempo-partitioning experiments. The comparison of activity coefficients and the phase boundary in DMPC/cholesterol mixtures with those of POPC/cholesterol mixtures suggests that acyl chain unsaturation leads to poorer mixing of cholesterol in phosphatidylcholine model membranes at higher temperatures (i.e., >35°C). In ternary solutions of DMPC, POPC, and cholesterol, it is found that cholesterol shows less deviation from ideality than in either of the two binary mixtures, and this implies that the phase separation occurs at higher cholesterol concentration than in either of the two binary mixtures. The present analysis suggests that there may not be a critical point in DMPC/cholesterol mixtures, even though phase separation does occur.
|
|
|
Heisenberg Spin Exchange and Molecular Diffusion in Liquid Crystals
A. Nayeem, S.B. Rananavare, V.S.S. Sastry, and Jack H. Freed.
J. Chem. Phys. 91, 6887-6905 (1989).
<doi: 10.1063/1.457358>
Publication #183
|
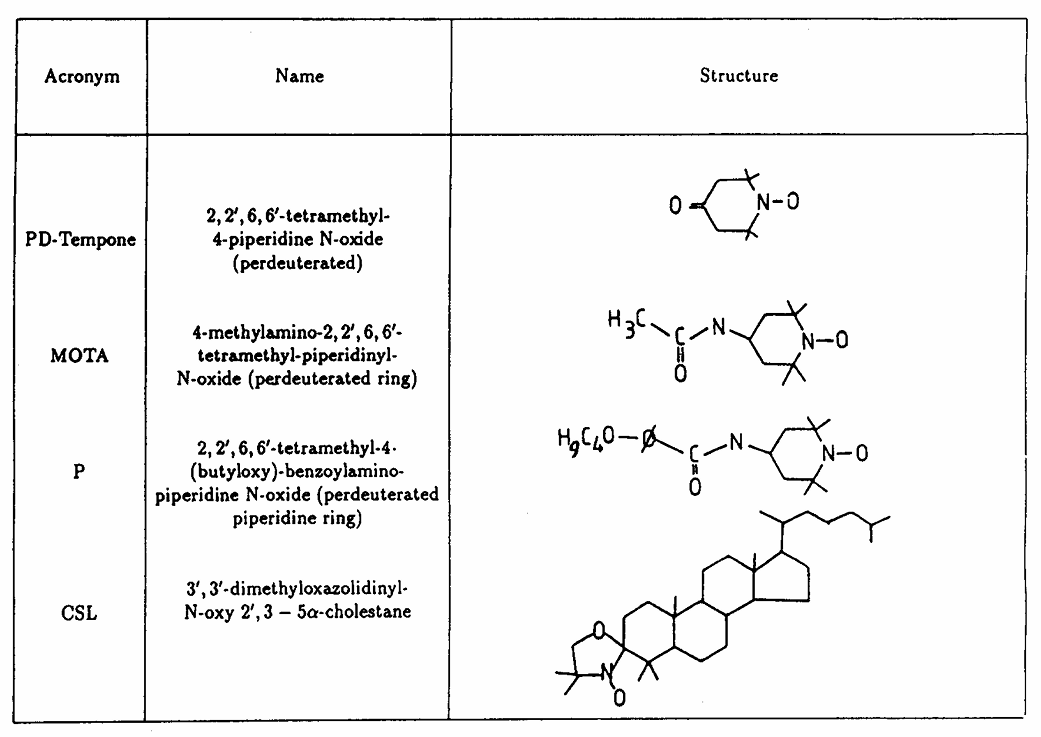
|
|
ABSTRACT: Heisenberg spin exchange (HE) studies of translational diffusion of the nitroxide radicals PD-tempone and P probe in two liquid crystalline solvents 6OCB–8OCB and 4O,6 are described. It is shown that while PD-tempone undergoes strong exchange in the two solvents, the more anisotropic P-probe exhibits a tendency toward weak exchange which becomes more prominent in the low temperature mesophases. The molecular diffusion rates measured from our HE studies are compared with rates measured over larger distances using electron-spin resonance (ESR) imaging methods; we find that, in similar thermotropic liquid crystals, the former are somewhat faster. Also, while diffusion rates for PD-tempone (using HE) in the ordered phases of 6OCB–8OCB are consistent with a single activation energy, those in 4O,6 show variations; suggesting that probe expulsion from core to chain regions in the former most likely occurs prior to SA formation, whereas in the latter it occurs in the SA phase. The absence of discontinuities in our diffusion data at the N–SA–RN transitions supports the belief that these transitions are subtle, with nothing dramatic occurring as the reentrant nematic (RN) phase is formed. The effect of including a potential of mean force U(r) between colliding radicals due to the liquid-crystal structure, is also considered. Our analyses indicate that the potential is of a repulsive nature [i.e., U(d)>0] suggesting the possibility of solvent molecules inhibiting collisions of radicals at distances shorter than the sum of their solvated radii. The influence of orientational ordering on HE involving nonspherical radicals is considered, but changes from strong to weak exchange in the ordered phases appear to depend on how τ1 the lifetime of the interacting radical pair is influenced by U(r). A careful effort is made to separate the HE effects from the intermolecular electron–electron dipolar (EED) interactions. It is suggested that anomalies in D obtained from HE vs EED in this and earlier studies may also be rationalized in terms of the effects of U(r).
|
|
|
Two-Dimensional Electron Spin Resonance
J. Gorcester, G.L. Millhauser, and J.H. Freed.
In Modern Pulsed and Continuous-Wave Electron Spin Resonance, L. Kevan and M. Bowman, Eds.
Wiley: NY, 1990; Chapter 3, pp. 119-194.
|
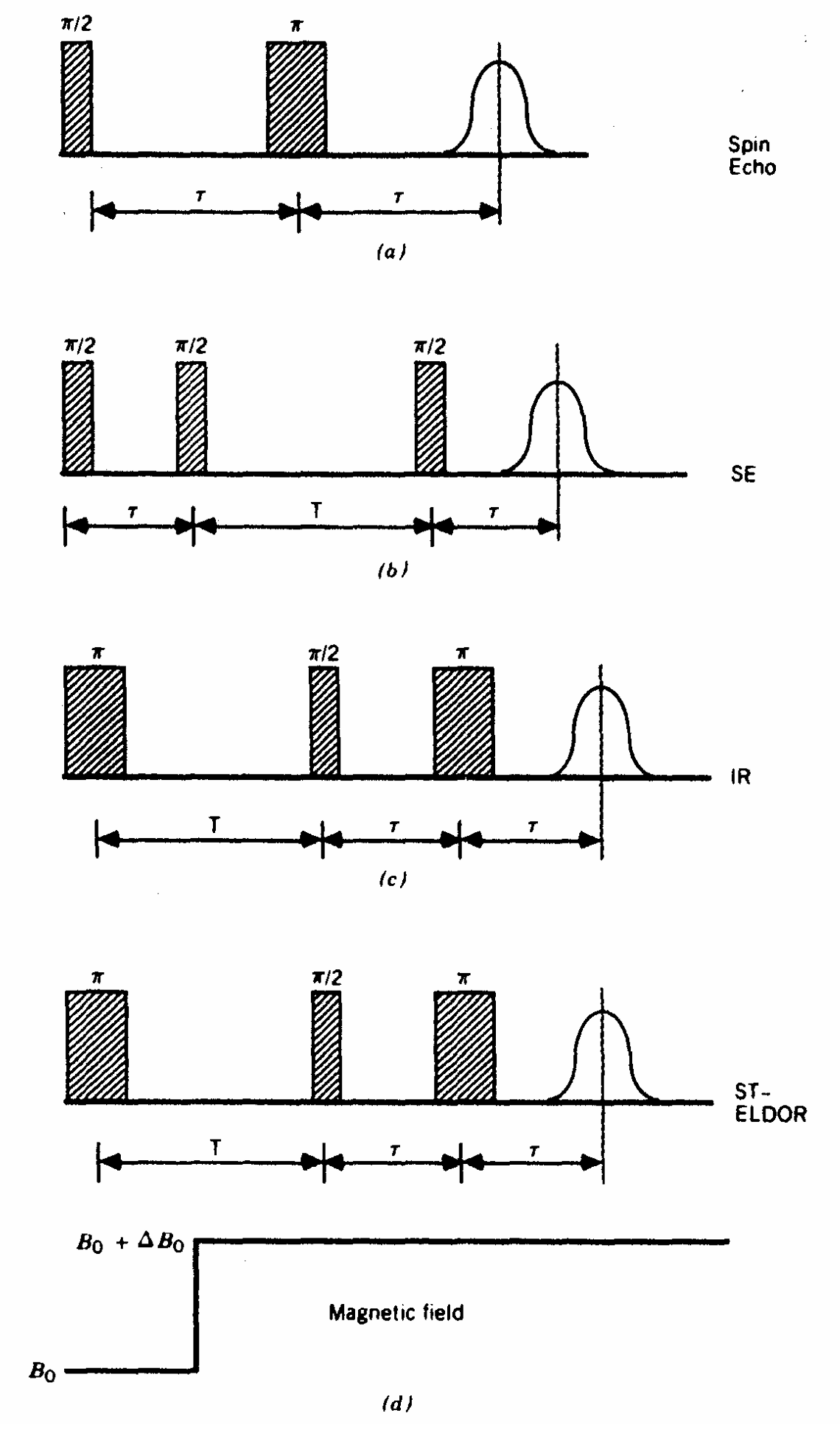
|
|
Two-Dimensional Electron Spin Resonance
J. Gorcester, G.L. Millhauser, and J.H. Freed.
In Modern Pulsed and Continuous-Wave Electron Spin Resonance, L. Kevan and M. Bowman, Eds.
Wiley: NY, 1990; Chapter 3, pp. 119-194.
Publication #182
|

|
|
INTRODUCTION: In 1979 one of us wrote a review in which a general formulation of time-domain ESR was described, and it was applied to saturation recovery ESR spectroscopy. In the same volume, Stillman and Schwartz provided a summary of the initial efforts in utilizing this formulation for ESR spin echoes and ENDOR spin echoes. Since that time there have been many experimental and theoretical developments on the subject of time-domain ESR, spin relaxation, and motional dynamics. We focus primarily (but not exclusively) in this chapter on the developments in time-domain ESR that pertain to studying molecular motions in condensed phases. Whereas the initial studies were based on spin-echo measurements of T2 (or TM, the phase-memory time) and T1, summarized in part in Section 2, the new developments have emphasized the advantages of two-dimensional spectroscopies that enable one to resolve more of the spectral and relaxation details that relate to motions.
These developments have occurred in two stages. Millhauser and Freed, realized that a conventional electron spin-echo (ESE) spectrometer could readily be modified to produce magnetic-field swept ESE decays, and then a single fast Fourier transformation (FFT) yields a 2D spectrum. The first type, described in Section 3, provides a mapping of the homogeneous line shapes across the inhomogeneous ESR spectrum, and it is very sensitive to the details of the motional dynamics. A second type, described in Section 4, provides a mapping of the cross-relaxation rates from each point in the spectrum and is also very informative of the motional dynamics.
|
|
|
Nuclear-Spin Waves in Polarized Atomic Hydrogen Gas: Temperature and Density Dependence in the Hydrodynamic and Knudsen Regimes
N.P. Bigelow, J.H. Freed, and D.M. Lee.
Phys. Rev. Lett. 63, 1609-1612 (1989).
<doi: 10.1103/PhysRevLett.63.1609>
PMID:
10040623
Publication #181
|
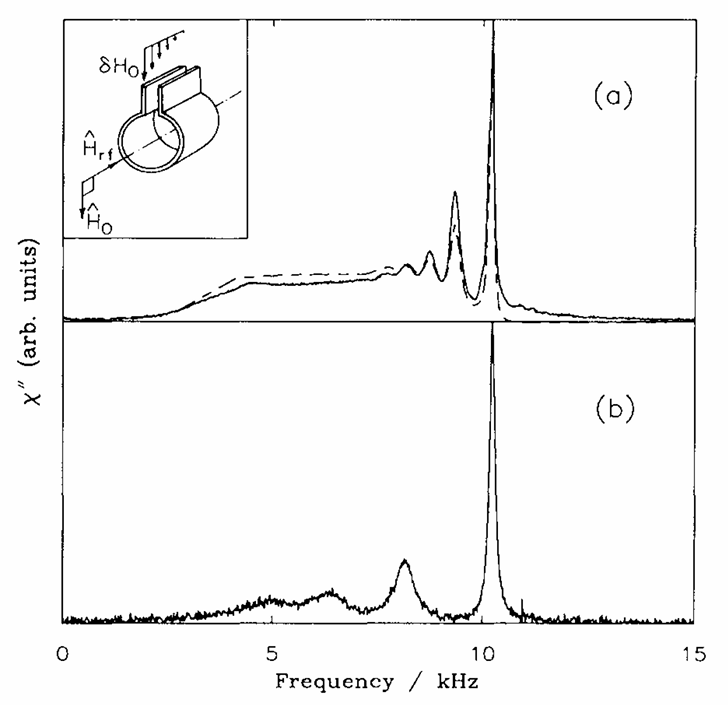
|
|
ABSTRACT: Accurate measurements on the nuclear-spin waves observed in the NMR spectrum of spin-polarized atomic hydrogen provide a quantitative test of theory. Results for temperatures between 160 and 537 mK and densities between 7×1015 and 5×1016 cm−3 display the expected magnitude and T−1∕2 divergence predicted for the spin-wave quality factor μ. It is shown that spin waves persist at low enough densities (Knudsen regime) that elastic wall collisions are more probable than atom-atom exchange collisions. An anomalous increase in spin-wave widths at low temperatures is explained in terms of the enhanced effects of sticking collisions of the atoms with the walls.
|
|
|
Discrete Microwave Absorption Lines in YBa2Cu3O7-δ Single Crystals
A. Dulčić, R.H. Crepeau, and J.H. Freed.
Physica C 160, 223-226 (1989).
<doi: 10.1016/0921-4534(89)90053-1>
Publication #180
|
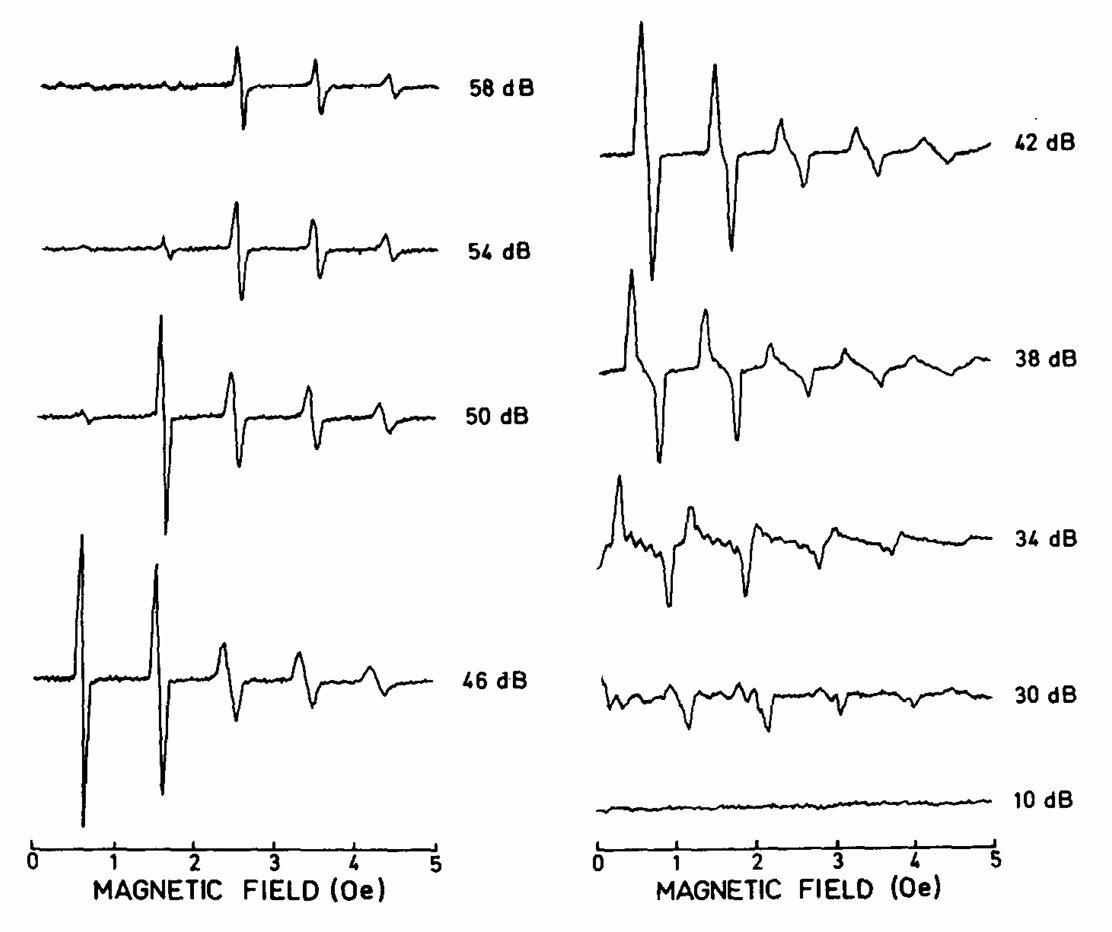
|
|
ABSTRACT: Discrete microwave absorption lines in YBa2Cu3O7−δ single crystals are analyzed as a function of microwave power, temperature and prior exposure to higher fields. The intensity of the low field lines shows critical dependence on the microwave power and temperature. The lines broaden and saturate with increasing microwave power. No broadening is observed at higher temperatures, but only a decrease in intensity. Exposure of the sample to progressively higher fields initially introduces a shift in the position of the lines dependent on the direction of the field scan, and then vanishing of the signal.
|
|
|
Electron Paramagnetic Resonance at 1 Millimeter Wavelengths
D.E. Budil, K.A. Earle, W.B. Lynch, and J.H. Freed.
In Advanced EPR: Applications in Biology and Biochemistry, A.J. Hoff, Ed.
Elsevier: Amsterdam, 1989; Chapter 8, pp. 307-340.
<doi: 10.1016/B978-0-444-88050-5.50013-6>
Publication #179
|
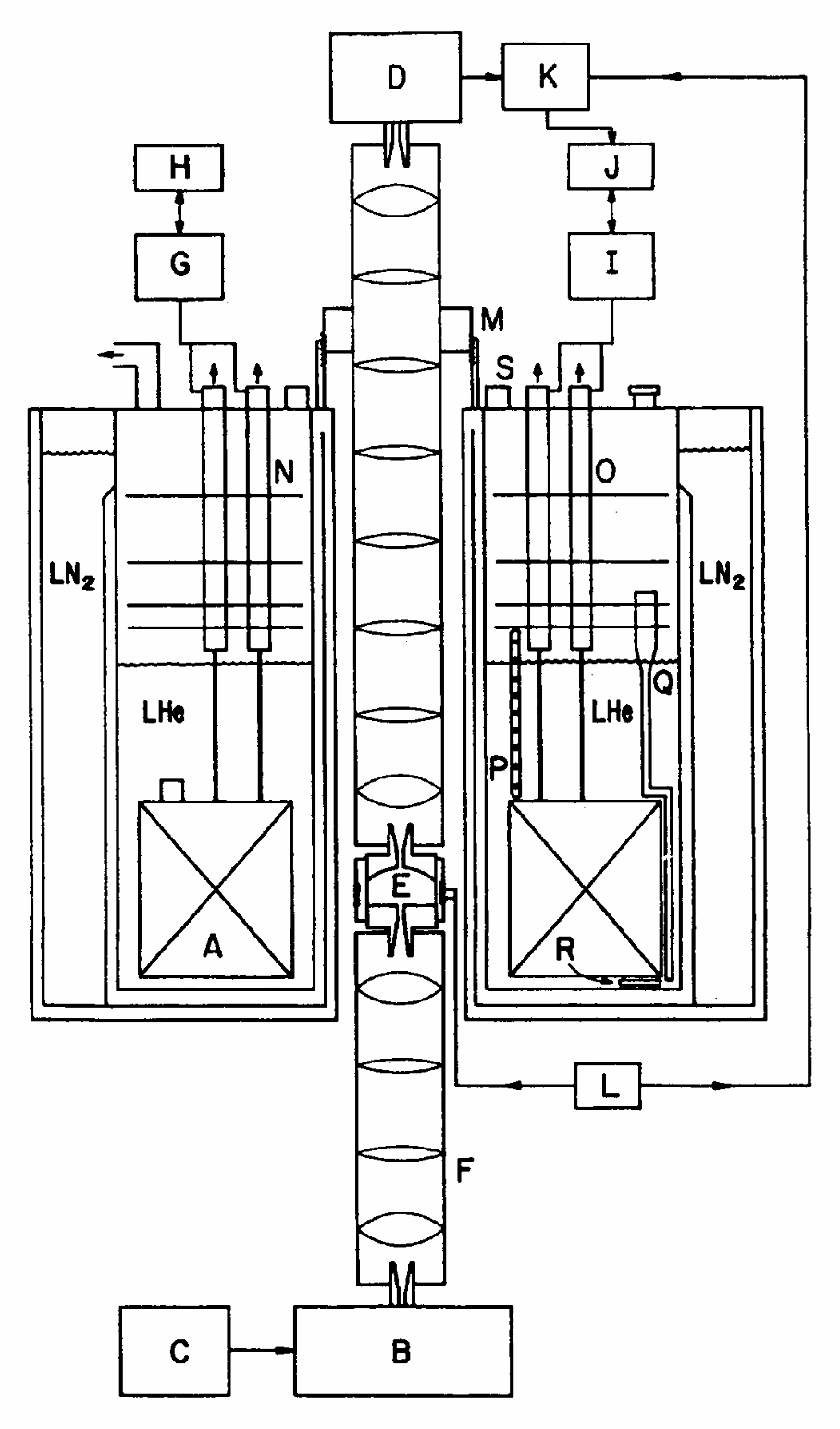
|
|
INTRODUCTION: For over two decades, new technical developments in EPR spectroscopy have lagged behind analogous advances in the field of NMR. Techniques such as Fourier-transform and two-dimensional correlation spectroscopy, which have long been standard in NMR, have only relatively recently found application in EPR. One of the most important advances in NMR has been the extension to high frequencies, requiring the use of superconducting magnets. This has provided large enhancements in sensitivity and spectral resolution, greatly broadening the utility of NMR techniques, especially for biological applications.
Recent years have witnessed a similar trend in EPR spectroscopy. The effort towards higher EPR fields and frequencies has in large part been fostered by an emerging technology in the synthesis and processing of signals in the far-infrared (FIR) and millimeter wave regions for applications in radar, communication, and radioastronomy. Notable in the field of EPR has been the development and application of a spectrometer working at λ &minus 2 mm (i.e. 148 GHz) and 5.3 Tesla fields by Lebedev and co-workers. They have reported EPR spectra of nitroxide spin labels in liquid and solid phases, emphasizing the significant advantages of high resolution for determining g-tensors and studying molecular dynamics in the slow-motional regime. The sensitivity at 148 GHz is also reported to be significantly better than that of conventional EPR spectrometers. More recently Möbius has constructed an EPR spectrometer operating at λ &minus 3.2 mm (94 GHz) in a 3.4 T field with similar advantages.
|
|
|
Two Dimensional and Fourier Transform ESR
J. Gorcester, G.L. Millhauser, and J.H. Freed.
In Advanced EPR: Applications in Biology and Biochemistry, A.J. Hoff, Ed.
Elsevier: Amsterdam, 1989; Chapter 5, pp. 177-242.
|
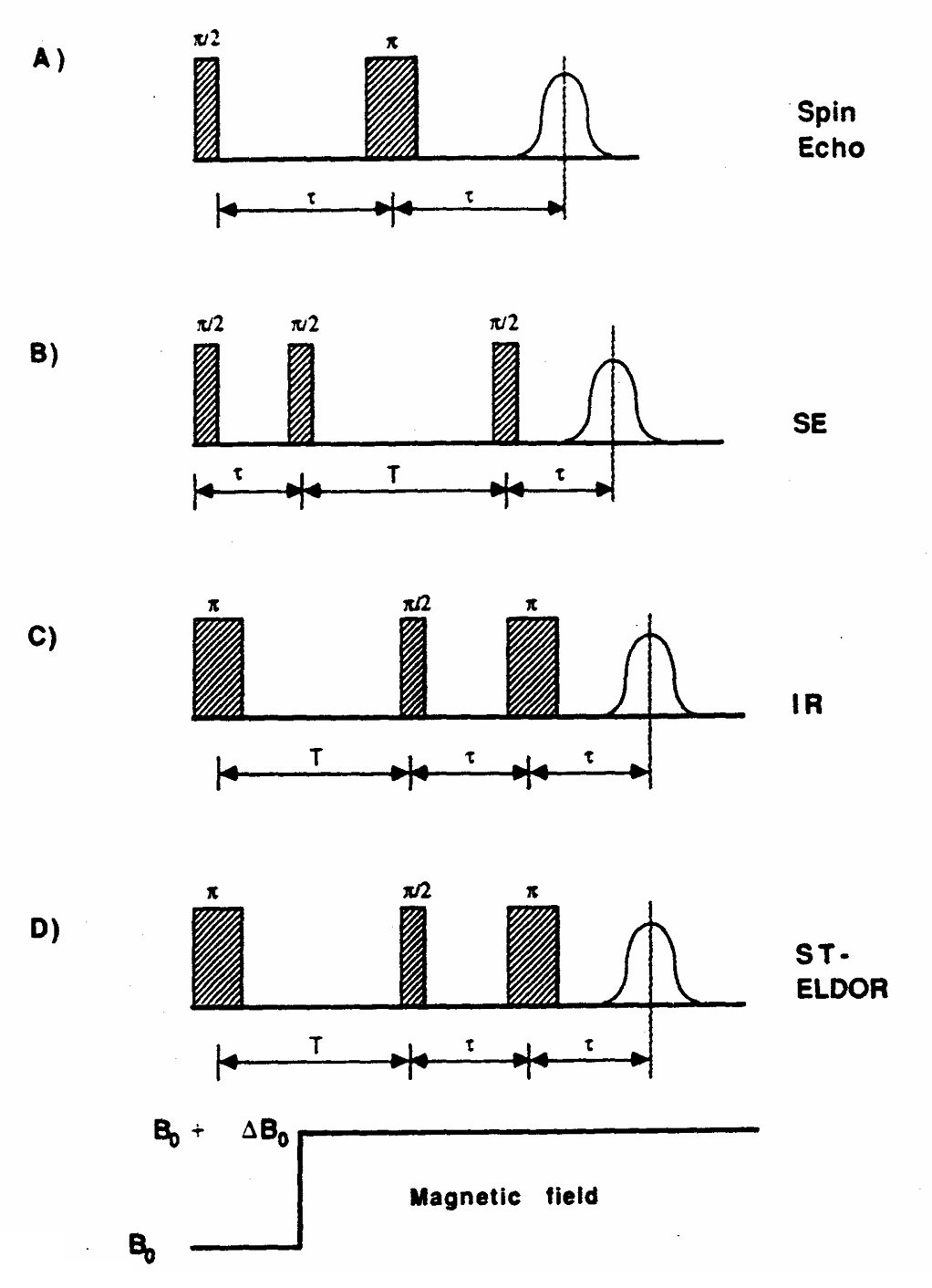
|
|
Two Dimensional and Fourier Transform ESR
J. Gorcester, G.L. Millhauser, and J.H. Freed.
In Advanced EPR: Applications in Biology and Biochemistry, A.J. Hoff, Ed.
Elsevier: Amsterdam, 1989; Chapter 5, pp. 177-242.
<doi: 10.1016/B978-0-444-88050-5.50010-0>
Publication #178
|

|
|
INTRODUCTION: In the past decade, an extensive range of modern time domain two-dimensional EPR techniques for the study of spin relaxation and motional dynamics of small and macromolecules in fluids, including membranes, have been developed based on electron-spin-echo and on Fourier Transform (FT) techniques. A primary concern has been with the spectra of nitroxide spin labels and spin probes.
The nitroxide spin-label technique has, in the last 20 years, permeated many areas of biophysics. The principal reliance has been on cw-EPR methods despite the somewhat limited features of the characteristic three-line hyperfine (hf) pattern from the 14N nucleus. Of course, as rotational motions slow down, spectra take on a greater range of shapes in the "slow-motional" regime. The analysis of such spectra is more complicated, but rewarding, because it can supply more detailed information on microscopic dynamics and ordering in membrane systems. Recent developments in computational methodology have now lead to a convenient but powerful computer program to simulate slow motional spectra that is readily available.
But the fact remains that these spectra exhibit rather low resolution to the details of the dynamics, a matter which is exacerbated by the proton (and∕or deuteron) super-hyperfine (shf) splittings. In a significant sense, EPR lagged behind NMR in its applications to the study of dynamic molecular structure, despite its virtues of greater sensitivity to fewer spins and the shorter time scale over which it examines motional dynamics. In NMR, after all, one can use spin echoes to measure homogeneous T2's; pulse techniques give T1's directly, and modern two-dimensional (2D) methods based on FT-NMR can provide detailed mappings of complicated cross-relaxation pathways.
|
|
|
Two-Dimensional FTESR and Solute Dynamics in Smectics
J. Gorcester, S.B. Rananavare, and J.H. Freed.
Bull. Magn. Reson. 11, 305-309 (1989).
Publication #177
|
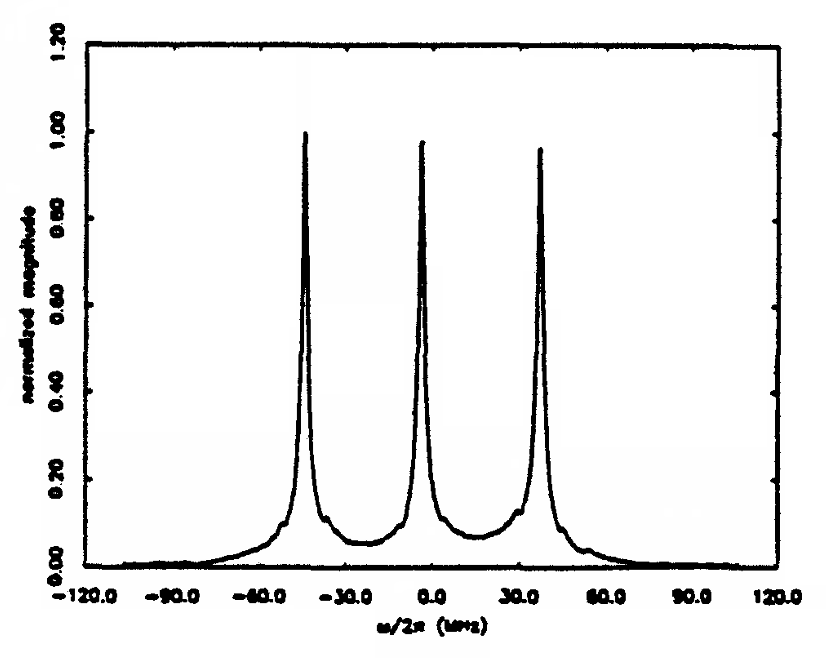
|
|
INTRODUCTION: Two-dimensional spectroscopies based on the Fourier transform technique may now be applied to the ESR of nitroxides. To achieve uniform coherent excitation of spectral bandwidths in excess of 90 MHz we have developed a TWT anode modulator with switching times of 3-4 ns. which is used in conjunction with two emitter coupled logic PIN switches connected in series. Simultaneous detection of in-phase and out-of-phase components of the time-domain response (free induction decay or FID) is achieved with a monolithic quadrature mixer and a two-channel digital storage oscilloscope. In order to correct for any phase or amplitude inaccuracy in the two detector channels, a four step phase alternation sequence is employed (CYCLOPS) using a 2-bit digital phase shifter capable of 7 ns. MW switching speeds for phase shifts of 0°, 90°, 180°, and 270°. To date we have obtained 15 Gauss of H1 in a bridged loop-gap resonator with a loaded Q of ˜ 35, enabling uniform excitation exceeding 90MHz bandwidths in single pulse (cf. Fig. 1) and multiple pulse (two-dimensional) FTESR experiments.
A two-dimensional correlation spectrum is obtained utilizing the three pulse sequence: π∕2 – t1 – π∕2 – tm – π∕2 – t2 by collecting the FID in the t2 domain with systematic variation of t1. The two-dimensional spectrum, obtained upon 2D FT of the motionally-narrowed nitroxide data, consists of cross-peaks connecting the auto-correlation lines appearing along the diagonal ω1 = ω2. A typical spectrum of this nature is shown in Fig. 2. The appearance of cross-peaks in these spectra indicates magnetization transfer (MT) induced by Heisenberg exchange (HE) and/or 14N spin relaxation during the mixing time tm, The rates of MT may be determined by measurement of cross-peak and auto-peak volume integrals, whereas the mechanism of MT may be deduced from the geometrical pattern of the spectral contours. These two mechanisms of MT may be distinguished due to the selection rule Δm1 = ±1 for 14N spin relaxation which restricts the cross-peaks to predominantly those connecting adjacent hyperfine (hf) lines. No such selection rule exists for the HE mechanism and thus HE contributes equally to all six cross-peaks. The quantitative accuracy of this method of determination of mechanisms and rates of MT has been confirmed by electron spin echo (ESE) experiments. We call this experiment 2D ELDOR because of its close similarity to that double resonance technique.
|
|
|
A Theoretical Model of Phospholipid Dynamics in Membranes
A. Ferrarini, P.L. Nordio, G.J. Moro, R.H. Crepeau, and J.H. Freed.
J. Chem. Phys. 91, 5707-5721 (1989).
<doi: 10.1063/1.457525>
Publication #176
|
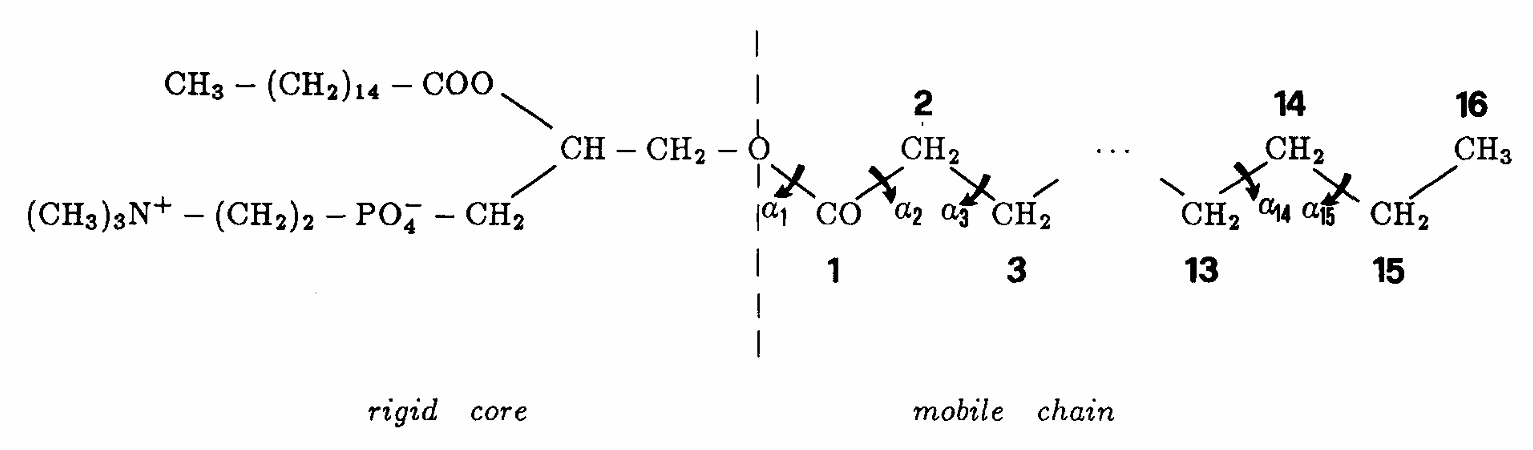
|
|
ABSTRACT: Static and dynamic properties related to the internal configurational motions have been calculated for the alkyl chains of phospholipid molecules in a membrane environment in the liquid crystal phase. The calculations have been performed for the chain 1 of 1,2-dipalmitoyl 3-sn-phosphatidylcholine (DPPC), a typical constituent of phospholipid membranes. Under the assumption of fixed bond lengths and bond angles, the internal dynamics of the chain is described in terms of 15 dihedral angles. The time evolution of the angular variables is assumed to be diffusional in character, and a master equation for transitions among the stable conformers is constructed from the energetics and hydrodynamics of the chain. This method is an extension to the time domain of the rotational isomeric state (RIS) approximation, which has been widely used to compute static properties of the chains. After calculation of the suitable correlation functions, effective rate constants relevant for spectroscopic and kinetic observables have been computed, and the results have been compared with those obtained by recent Brownian dynamics (BD) calculations. The position dependence of the rate constants along the chain has been examined with special reference to understanding the effects resulting from cooperativity in the conformational transitions. The overall spinning and tumbling of the chain has also been described by a diffusive model. The calculated spectral densities for the composite motional process have been used to rationalize the behavior of the relaxation times T1, T2, and T1ρ measured in deuterium nuclear magnetic resonance (NMR) experiments.
|
|
|
Composite Pulses in Time-Domain ESR
R.H. Crepeau, A. Dulčić, J. Gorcester, T.R. Saarinen, and J.H. Freed.
J. Magn. Reson. 84, 184-190 (1989).
<doi: 10.1016/0022-2364(89)90017-6>
Publication #175
|
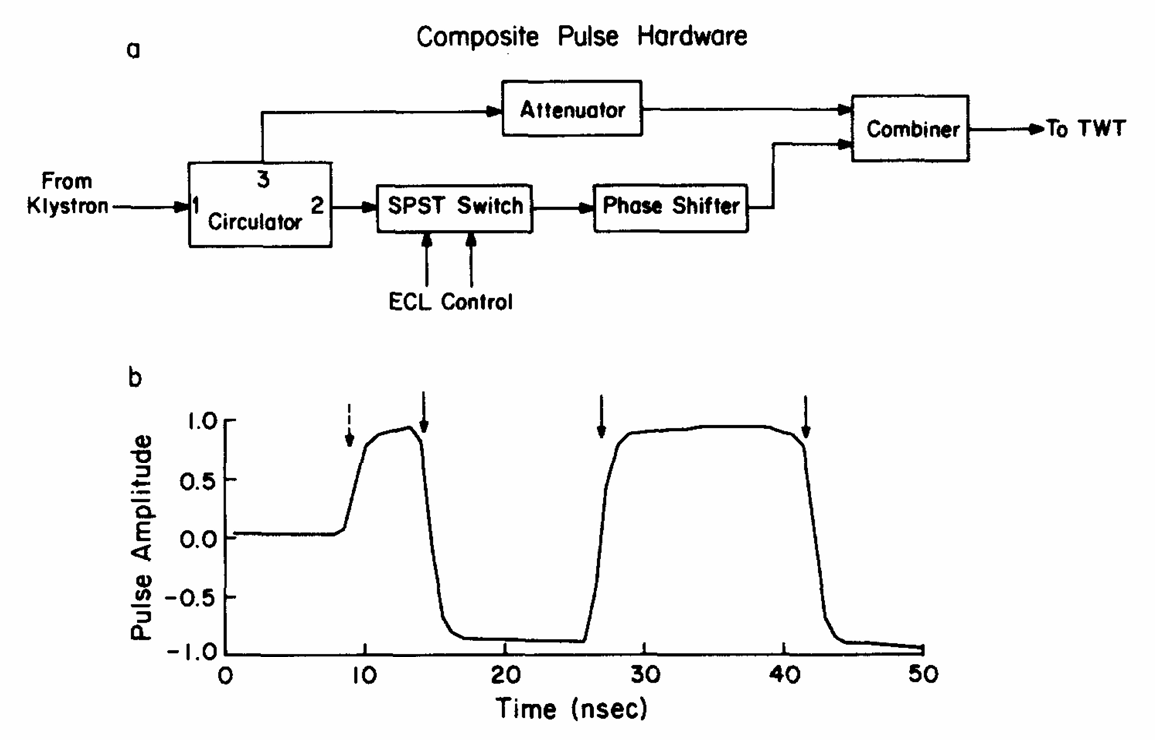
|
|
INTRODUCTION: Recently, new and important Fourier transform and two-dimensional Fourier transform ESR techniques have been developed which have the promise of revolutionizing ESR as they have NMR. The techniques have been described in detail and have been successfully applied to the class of nitroxide ESR spectra. A fundamental problem is the competing requirements of (1) delivering a large enough rotating microwave B1 field at the sample to irradiate the whole spectrum (i.e., 2γeB1 ≥ Δωs, where Δωs is the spectral extent) and at the same time (2) having a resonator with a low enough QL (loaded Q) that the short pulse of width tp required for a π∕2 (or π) rotation ofthe spins (i.e., γeB1tp = π∕2) is not significantly distorted. That is, we require that the half-power full bandwidth of the resonator, Δω = ω∕QL, obey Δω 3≥ Δωs. The problem, of course, is that for a given power, P0 incident on the resonator, B1 and QL are intimately related by P0 = μ0VcB21ν∕2QL, where ν is the resonator frequency and Vc is the effective volume.
From the above comments we can then state that the power required to irradiate the spectrum obeys P0 ∝ Δωs3 (3). Gorcester and Freed (GF) found for their 2D FT ESR spectrometer with a cavity resonator and TWT output of 1 kW that the optimum bandwidth was achieved with a B1 of 6 G (tp = 14 ns for a π∕2 pulse) and a QL ≈ 100 for which Δω∕2π = 95 MHz, corresponding to uniform spectral rotation over a bandwidth of about 34 MHz (= γe B1 ∕ π ). Nevertheless, they succeeded in obtaining wideband excitation for a motionally narrowed nitroxide with spectral extent, Δωs = 90 MHz. However, the single-pulse FID leads to an FT spectrum with outer hyperfine lines of reduced intensity (i.e., 50-60% of the central line). In a three-pulse 2D FT spectrum (i.e., 2D ELDOR), this attenuation of the outer peaks goes approximately as the cube of the attenuation of the simple FT spectral line. While appropriate corrections may be made for this effect in the analysis of the experiment, it does lead to reduced sensitivity.
|
|
|
ESR Study of the Dynamic Molecular Structure of a Reentrant Nematic Liquid Crystal
A. Nayeem and J.H. Freed.
J. Phys. Chem. 93, 6539-6550 (1989).
<doi: 10.1021/j100354a051>
Publication #174
|

|
|
ABSTRACT: ESR studies of anisotropic ordering and molecular dynamics using a variety of spin probes (PD-Tempone, MOTA, P-probe, and CSL) in a reentrant nematic (RN) liquid crystal mixture, 6OCB-8OCB, are described. In order to discern possible differences in the molecular behavior of reentrant mesogens, our results are compared with similar studies in those liquid crystals that, in spite of being structurally similar to 6OCB-8OCB (e.g., 8CB and S2, which exhibit bilayer smectic-A (SA) phases), do not exhibit reentrant behavior. Such comparisons show the following. (i) The SA-RN transition is very similar to the (normal) N-SA transition. (ii) The orientational ordering of P in the SA and RN phases of 6OCB-8OCB is consistent with more random packing of the chains than is observed in S2. It is suggested that the collective packing of the chains, which enhances the stability of the SA phase in a normal liquid crystal (e.g., S2), is frustrated in a reentrant nematic liquid crystal. (iii) The ordering of CSL, which packs with the cores, increases smoothly across the N-SA-RN transitions, showing that the packing of the aromatic cores is not sensitive to these phase transitions. In general, there are no dramatic changes in the dynamics of the probes upon entering the RN phase of 6OCB-8OCB, supporting the belief that the effects driving reentrance are very subtle. However, the reorientational dynamics of the P-probe indicate enhanced packing of the chains in the RN phase.
|
|
|
Dynamic Imaging of Diffusion by ESR
J.K. Moscicki, Y.K. Shin, and J.H. Freed.
J. Magn. Reson. 84, 554-572 (1989).
<doi: 10.1016/0022-2364(89)90120-0>
Publication #173
|
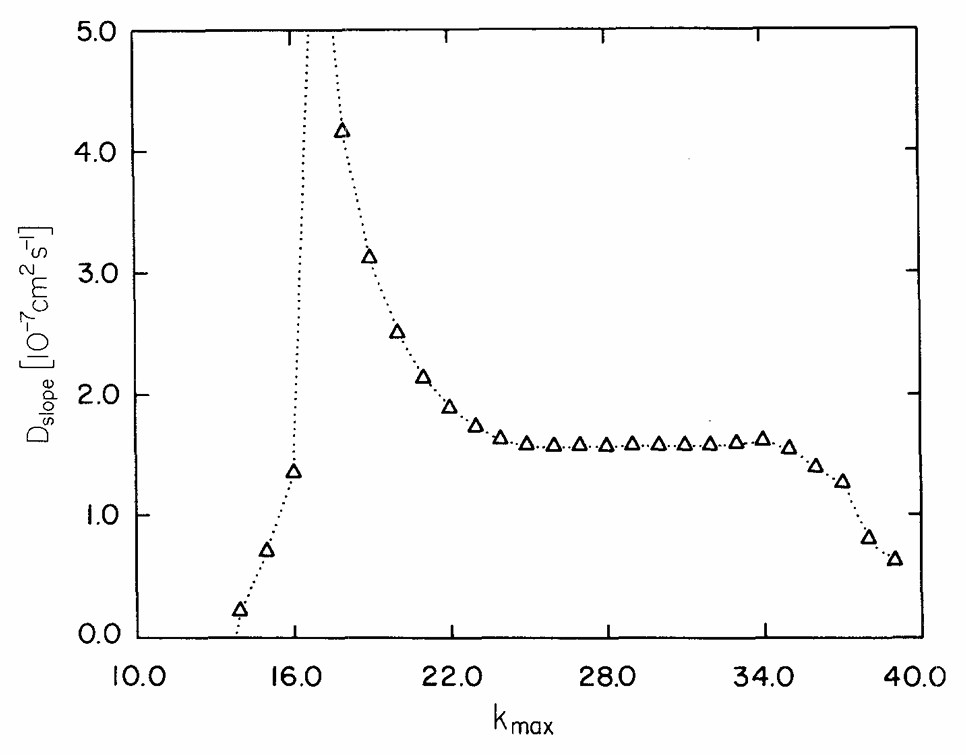
|
|
ABSTRACT: A comprehensive analysis of the accuracy and reliability of dynamic imaging of diffusion by ESR is presented. The importance of analyzing the data in Fourier space is emphasized, and a new method which enables the determination of the important Fourier modes while also providing a test of the reliability of the measurement of the diffusion coefficient, Dx, is presented. It is shown that values of Dx ∼ 10−9 cm2 s−1 can be measured in about one hour to 10–20% accuracy, whereas for Dx ∼ 10−7 cm2 s−1 the error should be below 1%. These statements are applicable for experiments in which there is unrestricted diffusion, and an initial Gaussian concentration profile. Systematic error resulting from non-Gaussian concentration profiles is shown to be relatively unimportant. However, the finite sweep time through a spectrum is found to yield a systematic error analogous to a Doppler shift, which tends to cancel only for the case of unrestricted diffusion. The experimental geometry in which there is diffusion from a reflective boundary requires special adjustments to align the gradient-on and -off spectra. A convenient and reliable method to accomplish this is presented. Nevertheless, there are inherently greater sources of error for this geometry, and this is confirmed by the experimental results. Utilizing the reflective boundary geometry, the longitudinal and transverse diffusion coefficients for PD-TEMPONE (15N labeled) in the nematic phase of MBBA at 20°C are 3.7 × 10−7 and 2.5 × 10−7 cm2 s−1, respectively. Their ratio of 1.5 is consistent with results for other probe molecules in MBBA.
|
|
|
Two-Dimensional Electron-Electron Double Resonance and Electron Spin-Echo Study of Solute Dynamics in Smectics
J. Gorcester, S.B. Rananavare, and J.H. Freed.
J. Chem. Phys. 90, 5764-5786 (1989).
<doi: 10.1063/1.456385>
Publication #172
|
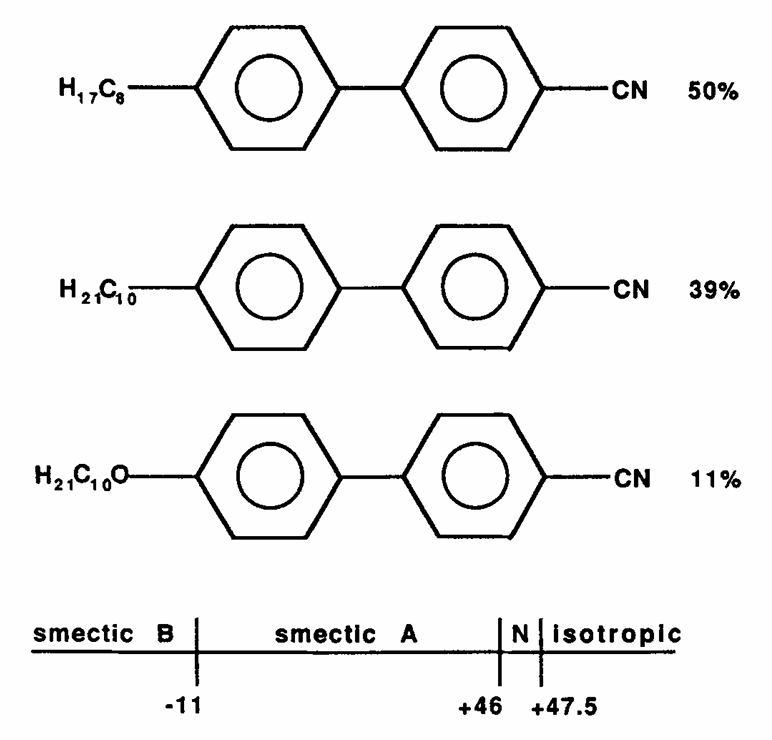
|
|
ABSTRACT: Electron spin–echo (ESE) and two-dimensional electron–electron double resonance (2D ELDOR) experiments have been performed as a function of director orientation and temperature in the smectic A phase of the liquid crystal S2 for the spin–probe PD-tempone(2×10−3 M). Over the entire temperature range studied (288–323 K) we observe significant 2D ELDOR cross peaks only for ΔMI=±1 indicative of 14N spin–relaxation and negligible Heisenberg exchange. From the angular dependent 14N spin–relaxation rates we obtain the dipolar spectral densities at the hyperfine (hf) frequency, whereas from a combination of ESE and 2D ELDOR we obtain the dipolar and Zeeman-dipolar spectral densities at zero frequency. The angular dependent spectral densities were successfully decomposed into their basic components in accordance with theory. The angular dependent spectral densities at the hf frequency are not predicted by a model of anisotropic rotational diffusion in a nematic orienting potential, but are consistent with predictions of a model due to Moro and Nordio of solute rototranslational diffusion in a McMillan-type potential. The angular dependence also indicates that order director fluctuations in the smectic phase are suppressed at frequencies on the order of 10 MHz. An additional contribution to solute reorientation due to cooperative hydrocarbon chain fluctuations is suggested to account for the behavior of the observed spectral densities at zero frequency. An evaluation of the relevance of several other dynamical models to this experimental work is also presented.
|
|
|
Berry's Geometrical Phases in ESR in the Presence of a Stochastic Process
D. Gamliel and J.H. Freed.
Phys. Rev. A 39, 3238-3255 (1989).
<doi: 10.1103/PhysRevA.39.3238>
PMID:
9901624
Publication #171
|
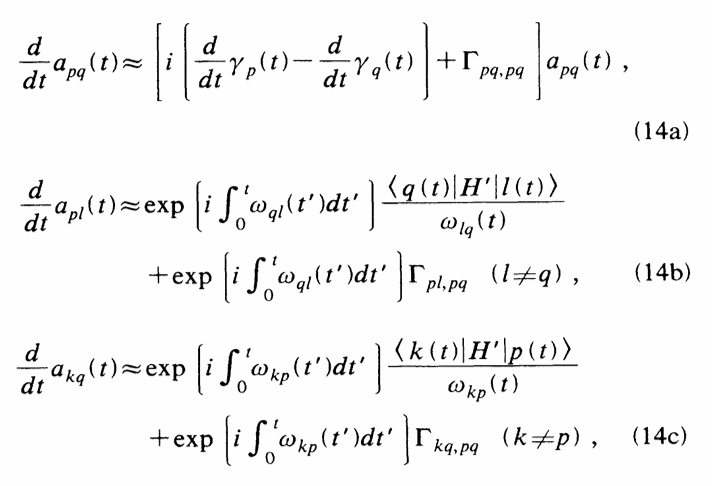
|
|
ABSTRACT: Berry's [Proc. R. Soc. London, Ser. A 392, 45 (1984)] geometrical phase is discussed in the context of dissipative evolution of an interacting spin system, governed by the stochastic Liouville equation. An analytical treatment is given for a possible ESR experiment on an interacting electron-nucleus system, modulated by two-site jumps. Geometrical phases are shown to be relevant to systems of this type, when their Hamiltonian changes slowly with time. A method for obtaining higher-order corrections to the adiabatic approximation is demonstrated. It is found that if the jumps are slow relative to the rate of change of the Hamiltonian, their effect reduces to familiar line broadening, and the geometrical phases may be observed experimentally. Equations are also set up for a similar ESR experiment on an electron-nucleus system undergoing isotropic rotational diffusion, and a brief discussion of the equations follows.
|
|
|
Dynamic Imaging of Lateral Diffusion by Electron Spin Resonance and Study of Rotational Dynamics in Model Membranes. Effect of Cholesterol
Y.K. Shin and J.H. Freed.
Biophys. J. 55, 537-550 (1989).
<doi: 10.1016/S0006-3495(89)82847-4>
PMID:
2539210
PMCID:
PMC1330507
Publication #170
|
|
|
ABSTRACT: The effects of cholesterol on the dynamics and the structural properties of two different spin probes, the sterol type CSL and the phospholipid type 16-PC, in POPC∕cholesterol oriented multilayer model membranes were examined. Our results are consistent with a nonideal solution containing cholesterol-rich clusters created by the self association of cholesterol in POPC model membranes. The lateral diffusion coefficient D of the spin probes was measured over the temperature range of 15 to 60°C and over the concentration range of 0 to 30 mol % of cholesterol in the model membrane by the electron spin resonance (ESR) imaging method. The rotational diffusion coefficients (including R⊥) and the order parameter S were determined utilizing a nonlinear least square ESR spectral simulation method. D, R⊥ and S of CSL deviate considerably from linear dependence on mole percent cholesterol. The D of CSL was decreased by a factor of four at 15°C and a factor of two at 60°C for concentrations of cholesterol over 10 mol %, whereas those of 16-PC were hardly affected. Cholesterol decreased R⊥ by a factor of 10 at 30 mol % of cholesterol, but it increased slightly that of 16-PC. A significant increase of S for CSL due to the presence of cholesterol was observed. It is shown how the difference in variation of S for CSL vs. 16-PC with composition may be interpreted in terms of their respective activity coefficients, and how a single universal linear relation is obtained for the S of both probes in terms of a scaled temperature. Simple but general correlations of D and of R⊥ with S were also found, which aid in the interpretation of these diffusion coefficients.
|
|
|
|
|
Magnetic-Field-Dependent Microwave Properties of YBa2Cu3Ox Single Crystals
A. Dulčić, R.H. Crepeau, and J.H. Freed.
Phys. Rev. B 39, 4249-4257 (1989).
<doi: 10.1103/PhysRevB.39.4249>
PMID:
9948762
Publication #168
|
|
|
ABSTRACT: Magnetic-field-dependent microwave properties of YBa2Cu3Ox single crystals are investigated as a function of temperature, oxygen annealing, and modulation amplitude. Three distinct signals are observed as the temperature is decreased from Tc to 2.4 K. It is established that the relative intensities of the signals depend on the degree of oxygen annealing. For the best annealed samples, only the high-temperature (just below Tc) signal remains prominent. The low-temperature signals are shown to be related to an inhomogeneous oxygen content in incompletely annealed samples. The hysteresis width increases at lower temperatures, indicating larger flux trapping. The sample-rotation experiments demonstrate that the microwave absorption depends on the state of the magnetization of the sample. In experiments in which the modulation amplitude is varied, we find that the previously observed anomaly in the shape of the signal is due to a fourth type of signal which has looplike form and is very sensitive to field reversals.
|
|
|
Theory for Dynamic Lineshapes of Strongly Correlated Two-Spin Systems
N.P. Benetis, D.J. Schneider, and J.H. Freed.
J. Magn. Reson. 85, 275-293 (1989).
<doi: 10.1016/0022-2364(89)90143-1>
Publication #167
|
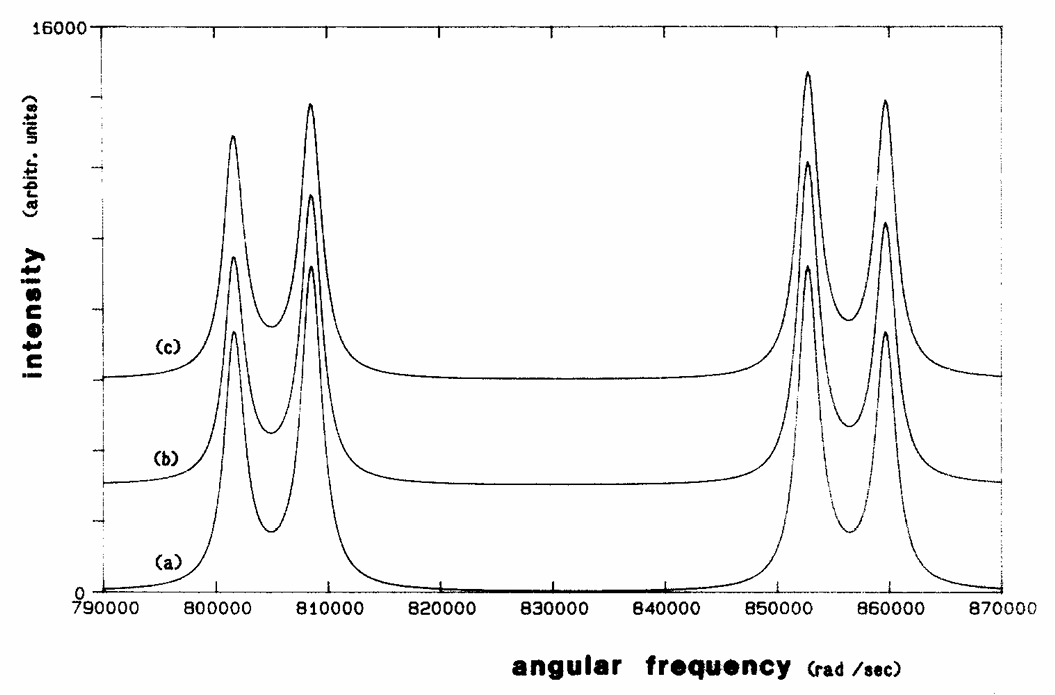
|
|
ABSTRACT: The calculation of spectra of two coupled spin systems within the stochastic Liouville equation formalism is systematized by the use of the coupled multipole-operator representation. The Liouville superoperator of the system is easily calculated in this representation in a unified way for all interactions by using a generalized Wigner-Eckart theorem in operator space. The spectrum is defined as the Fourier-Laplace transform of a single generalized correlation function, which, besides the autocorrelation of the observed spin can also contain contributions of the unobserved spin degrees of freedom, as well as all counterrotating parts when needed. The expression for the spectrum, which is an average of the appropriate first-order multipoles, is the same irrespective of the particular system; i.e., the multiplicity of the spin system, its states, and the relevant transition probabilities are all treated implicitly. This definition of the spectrum also introduces, through the preparation stage of the spin system prior to the free induction decay, another first-order multipole which is important for the case of strongly correlated spins. Two spins are strongly correlated when the difference in their Zeeman resonance frequency is comparable to their broadening. Accordingly, strong correlation is most likely to be important in low field and/or slow motion. The present method imposes no limitations on the relative strength of the relevant interactions or the rate of their modulation and is consequently valid in the slow-motion regime. A fast matrix-tridiagonalization method; based on the Lanczos algorithm, is used to calculate the spectrum, working directly in the frequency domain.
|
|
|
Studies of Nonlinear Spin Dynamics of Polarized Atomic Hydrogen Using NMR Pulses of Finite Duration in an Applied Magnetic Field Gradient
N.P. Bigelow, J.H. Freed, D.M. Lee, and B.W. Statt.
In Spin-Polarized Quantum Systems, S. Stringari, Ed.
World Scientific: New Jersey, 1989; pp. 227-230.
Publication #166
|
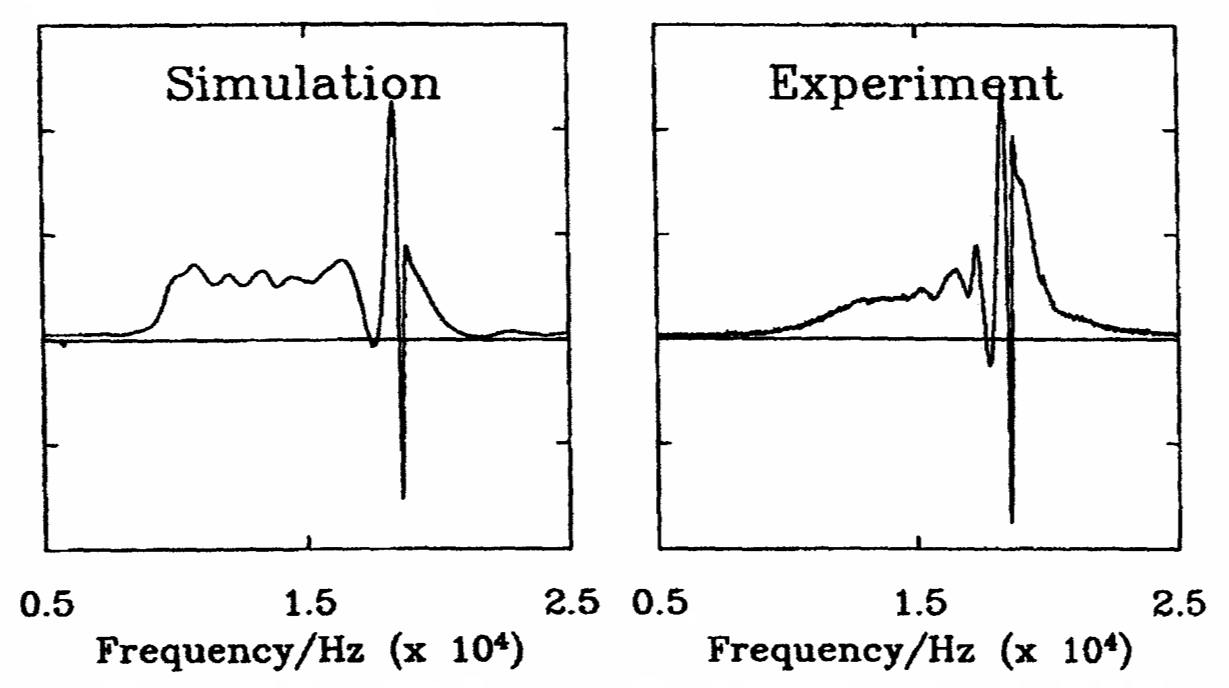
|
|
ABSTRACT: We describe the results of experiments on the effects of pulses of finite duration on the spin wave spectra of a sample of spin polarised atomic hydrogen in a static magnetic field gradient and for a range NMR tipping anglea. As the NMR pulae width is increased the spectral width of the pulse decreases and the spatial distribution of magnetization following the pulse is affected. Experiments have covered a range of spectral widths for the pulse from much narrower than to much wider than the inhomogeneous spectral width of the sample. Significant changes are observed in the Fourier transform spectra as a function of pulse length and will be compared to computer simulation and theory.
|
|
|
Time Domain Spectroscopy of Linear Spin Wave Modes of Polarized Atomic Hydrogen Using a Linear Least Squares Technique
N.P. Bigelow, J.H.Freed, D.M. Lee, and B.W. Statt.
In Spin-Polarized Quantum Systems, S. Stringari, Ed.
World Scientific: New Jersey, 1989; pp. 231-235.
Publication #165
|
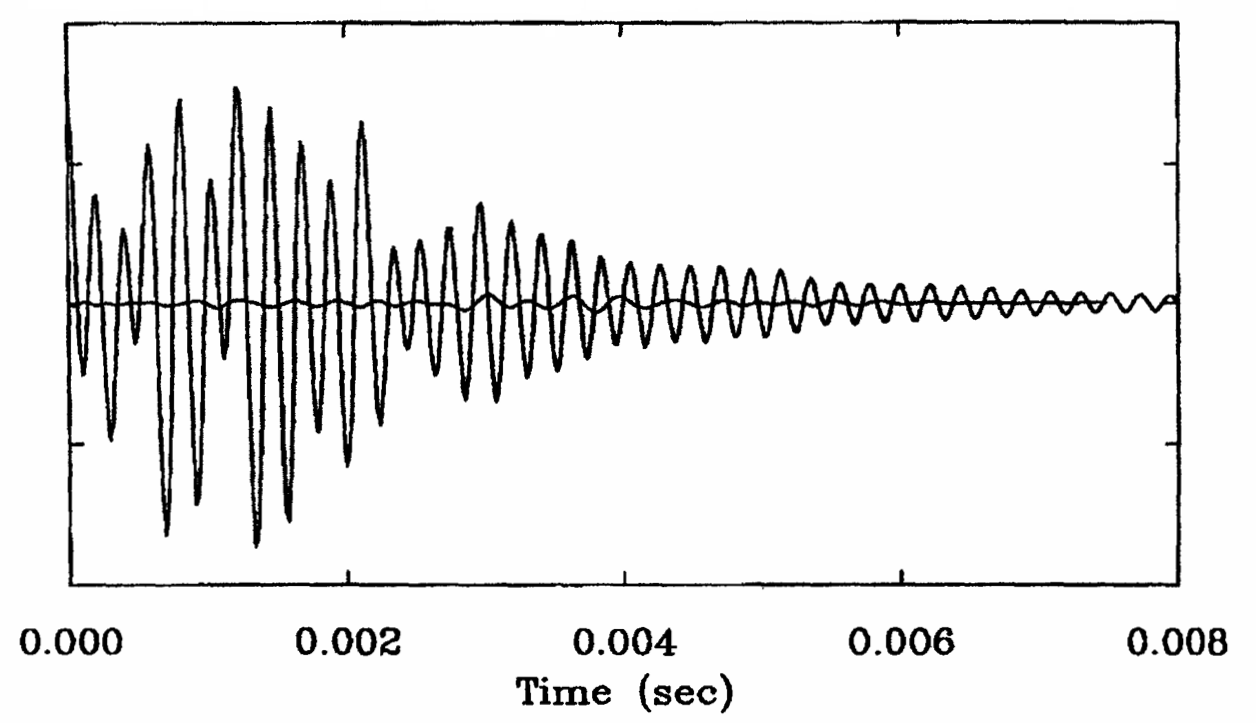
|
|
ABSTRACT: The Fourier transform spectra of the free induction decays from our small tipping angle pulsed NMR experiments in spin polarized atomic hydrogen are characterized by a number of sharp resonances superimposed on an inhomogeneously broadened lineshape. The complex nature of the spectra makes quantitative analysis of the individual spin wave modes difficult, in particular hindering the extraction of damping factors and relative phases. We describe an applicatian of a time domain fitting procedure based on a linear least squares analysis using linear prediction and singular value decomposition which yields numerical results that are compared directly to theoretical simulations.
|
|
|
Rapid Singular Value Decomposition for Time-Domain Analysis of Magnetic Resonance Signals by Use of the Lanczos Algorithm
G.L. Millhauser, A.A. Carter, D.J. Schneider, J.H. Freed, and R.E. Oswald.
J. Magn. Reson. 82, 150-155 (1989).
<doi: 10.1016/0022-2364(89)90175-3>
Publication #164
|
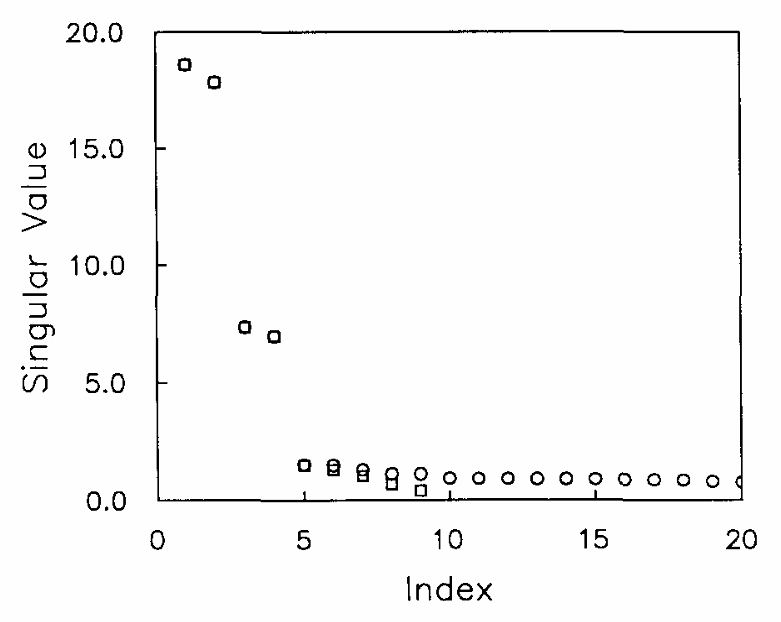
|
|
INTRODUCTION: Time-series analysis has become an integral part of signal interpretation in magnetic resonance and other experiments. As an example, linear prediction with singular value decomposition (LPSVD) and the related method, Hankel SVD (HSVD), have been recently applied to problems in NMR, 2D NMR, 2D electron spin-echo spectroscopy (2D ESE), Fourier transform ESR correlation spectroscopy, time-resolved femtosecond spectroscopy, and nonstationary neurological currents. The minimal goal of time-series analysis is to separate signal from noise. Spectral transform methods, such as FFT filtering, maximum entropy (MEM), and LPZ, replace the original data with a new time series (or its spectrum) that has an enhanced signal-to-noise ratio. In contrast spectral decomposition methods, such as LPSVD or HSVD, which assume a certain functional form for the time series, provide both a recipe for separating signal from noise and a listing of the harmonic components contained in the original data. This harmonic list simplifies spectral analysis and can be used to (1) extend the noise-reduced time series, (2) selectively reconstruct certain regions of the harmonic spectrum, and (3) data-compress the signal. The substantial benefits of spectral decomposition are often offset by the requirements of considerable computational time and memory storage. For a time series of N data points spectral transform techniques usually require computational time of O(N log2N) to O(N2) and storage of O(N) whereas spectral decomposition, such as HSVD, requires computational time of O(N3) and storage of O(N2).
We have investigated a new approach for solving the spectral decomposition problem which is based on the Lanczos algorithm (LA). In the past the LA has provided an effective means for solving slow-motional magnetic resonance spectra as well as other complicated problems in chemical physics. The LA is an extremely fast method for solving large eigenelement problems when either (1) all the eigenelements of a sparse matrix are required or (2) a small set of relevant eigenelements of a dense (or sparse) matrix of special structure are required. (A Lanczos-Prony method for time-series problems has been previously proposed, but it does not benefit from SVD as an intermediate step nor is it based on the LA that we employ.) The goal of this report is to discuss our recent findings on the applicability of the LA for spectral decomposition. We base our approach on the HSVD. The time-consuming singular value decomposition is performed with the LA and we find that the computational time for Lanczos-HSVD (LA-HSVD) is of O(N2) and the storage is of O(N).
|
|
|
Calculating Slow Motional Magnetic Resonance Spectra: A User's Guide
D.J. Schneider and J.H. Freed.
In Spin Labeling: Theory and Applications, Vol. III, L.J. Berliner and J. Reuben, Eds.
Biological Magnetic Resonance, Plenum: New York, 1989; Volume 8, pp. 1-76.
<doi: 10.1007/978-1-4613-0743-3_1>
Publication #163
|
|
|
INTRODUCTION: In the first volume of Spin Labeling: Theory and Applications a chapter was written by one of us (J.H.F.) in which a detailed theory for the interpretation of ESR spectra of spin labels in the slow motional regime was presented. The specific emphasis of that review was on the interpretation of nitroxide spin label spectra and contained many such examples. In the ensuing 13 years, there have been a number of important developments. First and foremost has been the development and implementation of powerful computational algorithms that have been specifically tailored for the solution of these types of problems. The use of these algorithms often leads to more than an order-of-magnitude reduction in computer time for the calculation of any given spectrum as well as a dramatic reduction in computer memory requirements. Concomitant with these improvements in computational methodology has been the revolution in the power and availability of computer hardware. Taken together, these improvements in hardware and software have made it possible to quickly and conveniently perform spectral calculations on small laboratory computers which formerly required the resources of a large mainframe computer. The increase in the available computing power has also made it feasible to incorporate more sophisticated models of molecular structure and dynamics into the line-shape calculation programs.
|
|
|
Microwave study of YBa2Cu3Ox single crystals
A. Dulčić, R. H. Crepeau, and J. H. Freed
Phys. Rev. B 38, 5002-5005 (1988).
<doi: 10.1103/PhysRevB.38.5002>
PMID:
9946899
Publication #162
|
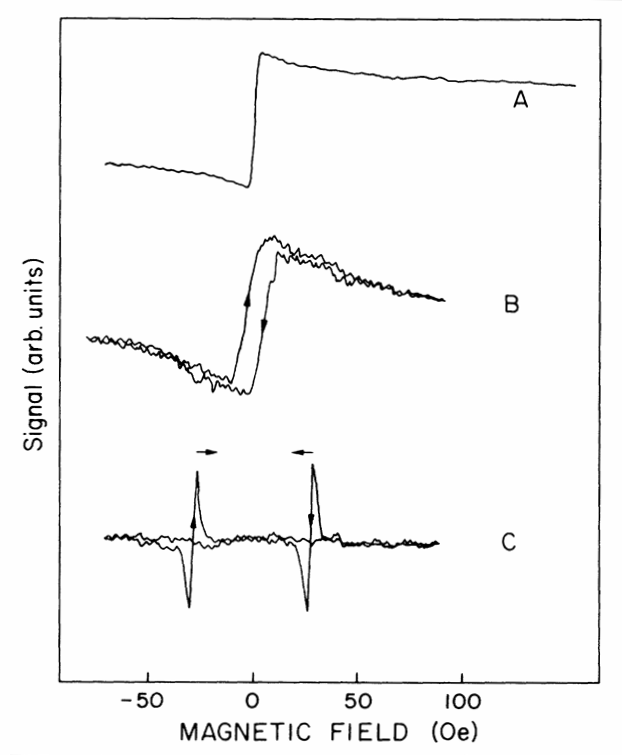
|
|
ABSTRACT: Absorption of microwaves (9.3 GHz) was measured as a function of an external applied magnetic field in single crystals of YBa2Cu3Ox. Three distinct types of signals were observed successively as the temperature was lowered from Tc to 2.4 K. They differ in widths, angular dependences, and hysteretic properties. We discuss possible microwave loss mechanisms and a model based on internal Josephson junctions to explain our observation.
|
|
|
1-mm wave ESR spectrometer
W.B. Lynch, K.A. Earle, and J.H. Freed.
Rev. Sci. Instr. 59, 1345-1351 (1988).
<doi: 10.1063/1.1139720>
Publication #161
|
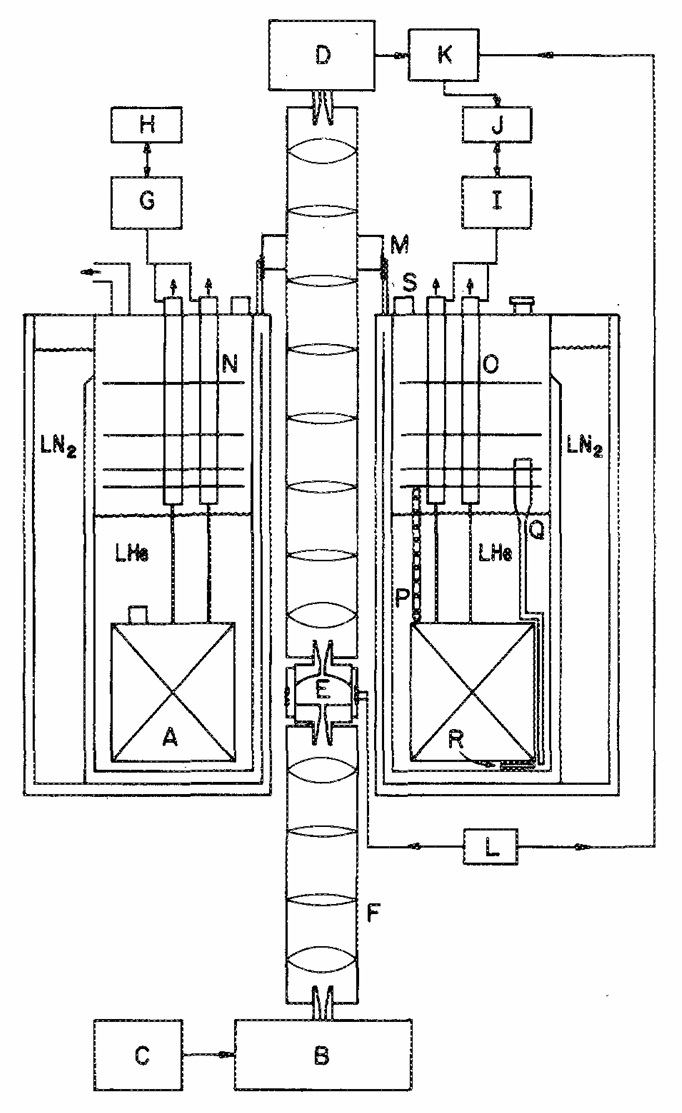
|
|
ABSTRACT: An ESR spectrometer operating at 250-GHz frequency (1.22-mm wavelength) and 9-T magnetic field is described. The utilization of far-infrared (FIR) technology greatly simplifies its design and performance. Good frequency and field stability are achieved by unique designs which also conveniently permit the magnetic field to be swept. The potential utility of FIR–ESR is illustrated with examples of spectra from polycrystalline and liquid samples. In the latter case the increased spectral sensitivity to motional dynamics is stressed. Several ways in which the FIR–ESR spectrometer may be improved are also discussed.
|
|
|
E.S.R. and D.S.C. Investigations of Phase Transitions in Polymorphic 4-n-Alkoxybenzylidene-4′-n-Alkylanilines
S.B. Rananavare, V.G.K.M. Pisipati, and J.H. Freed.
Liq. Cryst. 3, 957-976, (1988).
<doi: 10.1080/02678298808086552>
Publication #160
|
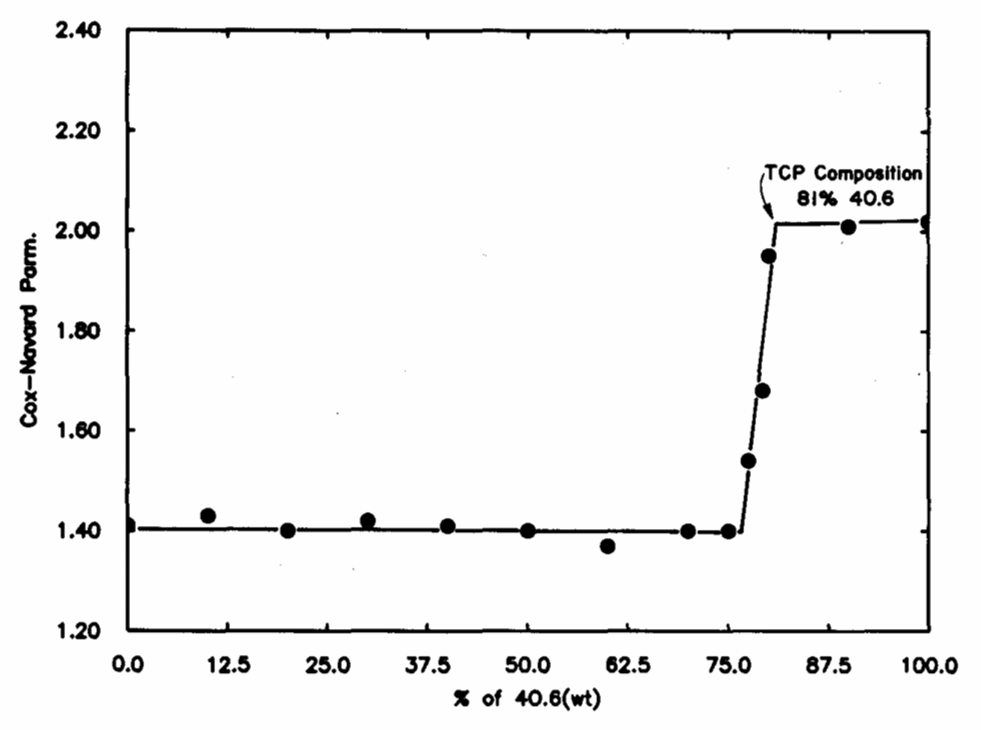
|
|
ABSTRACT: It is shown that the McMillan parameter M = TSAN∕TN1 (where TSAN and TNI are respectively the temperatures of the smectic A to nematic (SAN) and the nematic to isotropic (NI) phase transitions) is useful in analysing the crossover between second and first order behaviour of the SAN transition in the nO.m homologous liquid crystal series (the 4-n-alkoxybenzylidene-4′-n-alkylanilines). Using a phase diagram of orientational ordering versus M for this series, as obtained in this work (from E.S.R. and D.S.C), a symmetric tricritical point with mean field exponent β2 = 1 is demonstrated. In a preliminary study of E.S.R. linewidth parameters B and C of nitroxide spin probes dissolved in members of the nO.m series exhibiting a first order SAN transition, critical-type divergences are observed near this transition. In the case where M is closer to 0⋅959 (the value at the tricritical point), these divergences appear similar to those previously observed in related nO.m members with a second order SAN transition; however, they are considerably enhanced for an M value closer to unity (i.e. more removed from the tricritical point). This indicates the importance of coupling between orientational and positional order parameters in the observed critical-type divergences.
|
|
|
Rapid Determination of Translational Diffusion Coefficients Using ESR Imaging
D.A. Cleary, Y.K. Shin, D.J. Schneider, and J.H. Freed.
J. Magn. Reson. 79, 474-492 (1988).
<doi: 10.1016/0022-2364(88)90084-4>
Publication #159
|
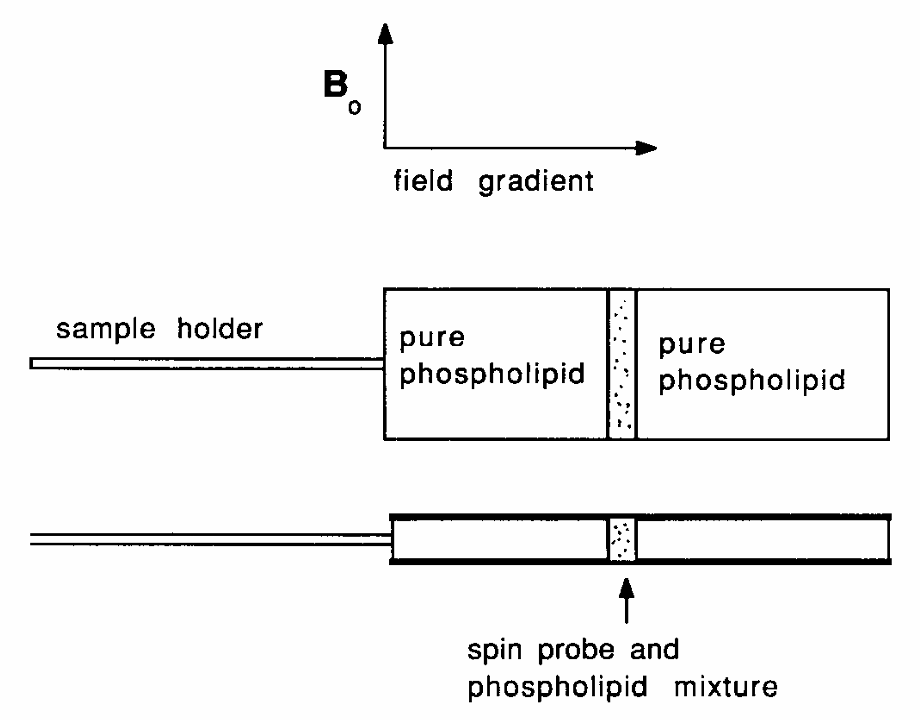
|
|
ABSTRACT: The use of ESR imaging for the rapid determination of diffusion coefficients, D, in anisotropic solvents is reported, and the use of Fourier transforms of the concentration profile to determine D is discussed. An approximate rapid analysis technique is also presented. The diffusion coefficients for motion perpendicular to the director axis for 4-oxo2,2,6,6,-tetramethylpiperidine-l-oxyl (TEMPONE) and octylbenzoyl spin label (OBSL) in the nematic phase of Phase V have been determined. They are D(TEMPONE) = 1.25 × 10−7 cm2 s-1 and D(OBSL) = 0.48 × 10−7 cm2 s-1 at 294 K. The lateral diffusion coefficients for cholestane spin label (CSL) in POPC and 16PC in DMPC were also determined. The diffusion coefficients were measured as a function of temperature from 11 to 60°C and they range from 4 × 10−8 to 2 × 10−7 cm2 s-1. Activation energies for diffusion in the two systems are Ea(CSL/POPC) = 6.30 kcal mol−1 and Ea(16PC/DMPC) = 8.63 kcal mol−1.
|
|
|
Linear Prediction and Projection of Pure Absorption Lineshapes in Two-Dimensional FTESR Correlation Spectroscopy
J. Gorcester and J.H. Freed.
J. Magn. Reson. 78, 292-301 (1988).
<doi: 10.1016/0022-2364(88)90272-7>
Publication #158
|
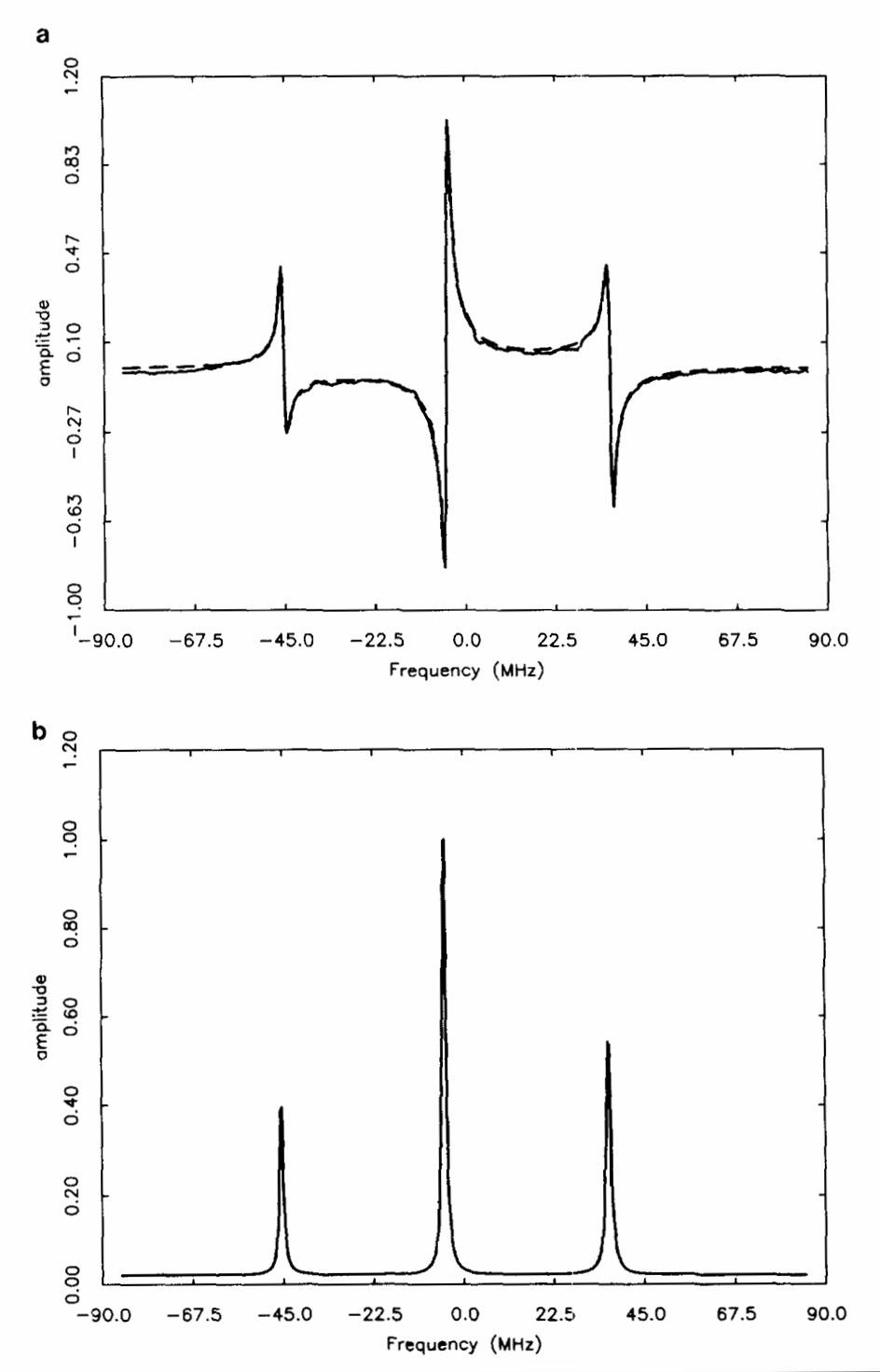
|
|
ABSTRACT: New ESR experiments based on techniques of two-dimensional correlation spectroscopy have been shown to be useful in the determination of magnetization transfer rates in motionally narrowed nitroxides. Spectral enhancement based upon linear prediction with singular value decomposition (LPSVD) is applied in the present work to project 2D absorption lineshapes and to dramatically improve the signal/noise ratio. Heisenberg spin exchange rates obtained from volume integrals of LPSVD-projected 2D absorption lineshapes compare well with those obtained from absolute value peak amplitudes in the case of 2,2,6,6-tetramethyl-4-piperidone-N-oxyl-d16 (pd-tempone) dissolved in toluene-d8 (where theory predicts that the two should agree).
|
|
|
Two-Dimensional Fourier Transform ESR Correlation Spectroscopy
J. Gorcester and J.H. Freed.
J. Chem. Phys. 88, 4678-4693 (1988).
<doi: 10.1063/1.453782>
Publication #157
|
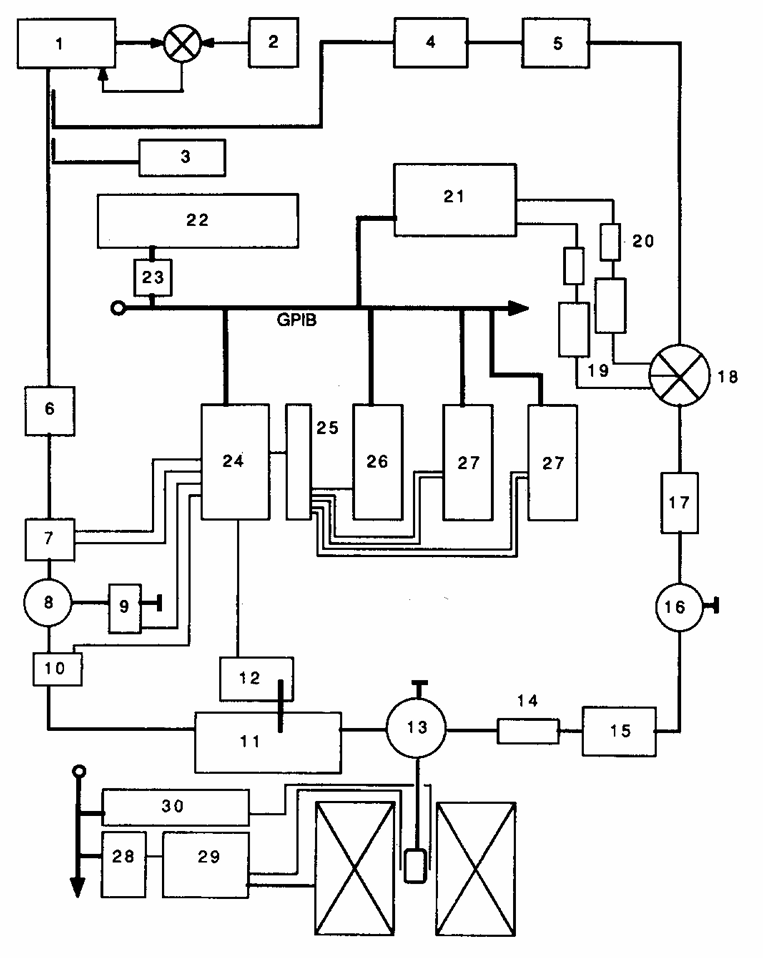
|
|
ABSTRACT: We describe our pulsed two-dimensional Fourier transform ESR experiment and demonstrate its applicabilty for the double resonance of motionally narrowed nitroxides. Multiple pulse irradiation of the entire nitroxide spectrum enables the correlation of two precessional periods, allowing observation of cross correlations between hyperfine lines introduced by magnetization transfer in the case of a three-pulse experiment (2D ELDOR), or coherence transfer in the case of a two-pulse experiment (COSY). Cross correlations are revealed by the presence of cross peaks which connect the autocorrelation lines appearing along the diagonal ω1=ω2. The amplitudes of these cross peaks are determined by the rates of magnetization transfer in the 2D ELDOR experiment. The density operator theory for the experiment is outlined and applied to the determination of Heisenberg exchange (HE) rates in 2,2,6,6-tetramethyl-4-piperidone-N-oxyl-d15 (PD-tempone) dissolved in toluene-d8. The quantitative accuracy of this experiment is established by comparison with the HE rate measured from the dependence of the spin echo T2 on nitroxide concentration.
|
|
|
|
|
Measurement of the Longitudinal Spin Diffusion Coefficient in Spin Polarized Atomic Hydrogen
N.P. Bigelow, J.S. Denker, B.W. Statt, D.M. Lee, and J.H. Freed.
Jap. J. of Appl. Phys. 26, 233-234 (1987).
<doi: 10.7567/JJAPS.26S3.233>
Publication #155
|
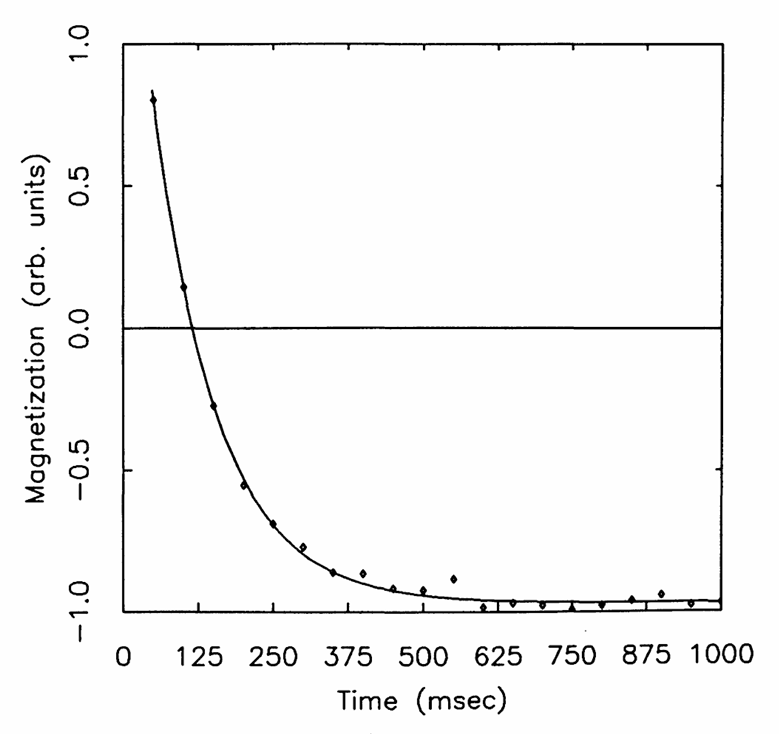
|
|
ABSTRACT: We report on the measurement of the longitudinal spin diffusion coefficient in doubly spin polarized atomic hydrogen. Measurements were made using pulsed nuclear magnetic resonance to probe the transport of magnetization between a sealed container and a large reservoir. Results are presented for densities between 1.4×1015 and 4.8×1016 cm-3 at temperatures between 246 and 470 mK in a magnetic field of 7.7 Tesla and are found to be in reasonable agreement with theoretical calculations by Lhuillier.
|
|
|
Nematic Order Near a Tricritical Nematic-Smectic A Phase Transition
S.B. Rananavare, V.G.K.M. Pisipati, and J.H. Freed.
Chem. Phys. Lett. 140, 255-262 (1987).
<doi: 10.1016/0009-2614(87)80453-0>
Publication #154
|
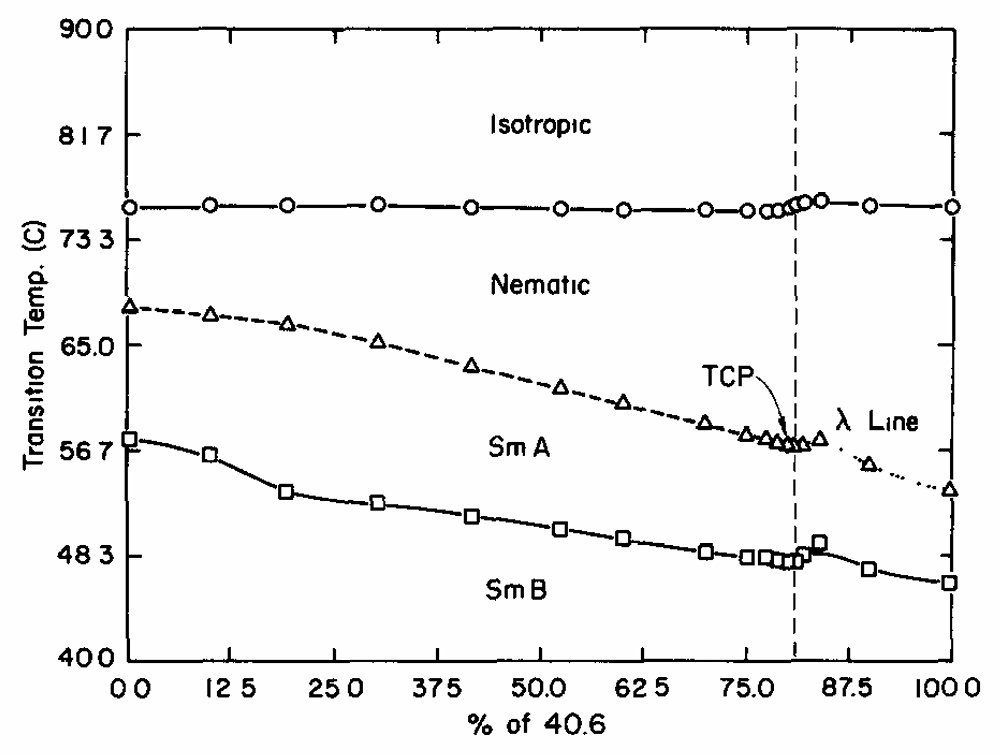
|
|
ABSTRACT: A symmetric tricritical point (TCP) is demonstrated for the nematic-smectic A transition by blending two nearly identical isomers and by showing that the difference in coexistence values of the nematic order parameter S goes to zero with TCP exponent β2 = 1 as TTCP is approached. A horizonta λ-line is associated with an observed universal behavior of S when a new scaling of T is used.
|
|
|
Spin Relaxation and Motional Dynamics
D.J. Schneider and J.H. Freed.
In Lasers, Molecules, and Methods, J.O. Hirschfelder, R.E. Wyatt, and R.D. Coalson, Eds.
Adv. Chem. Phys. 73, pp. 387-528 (1989).
|
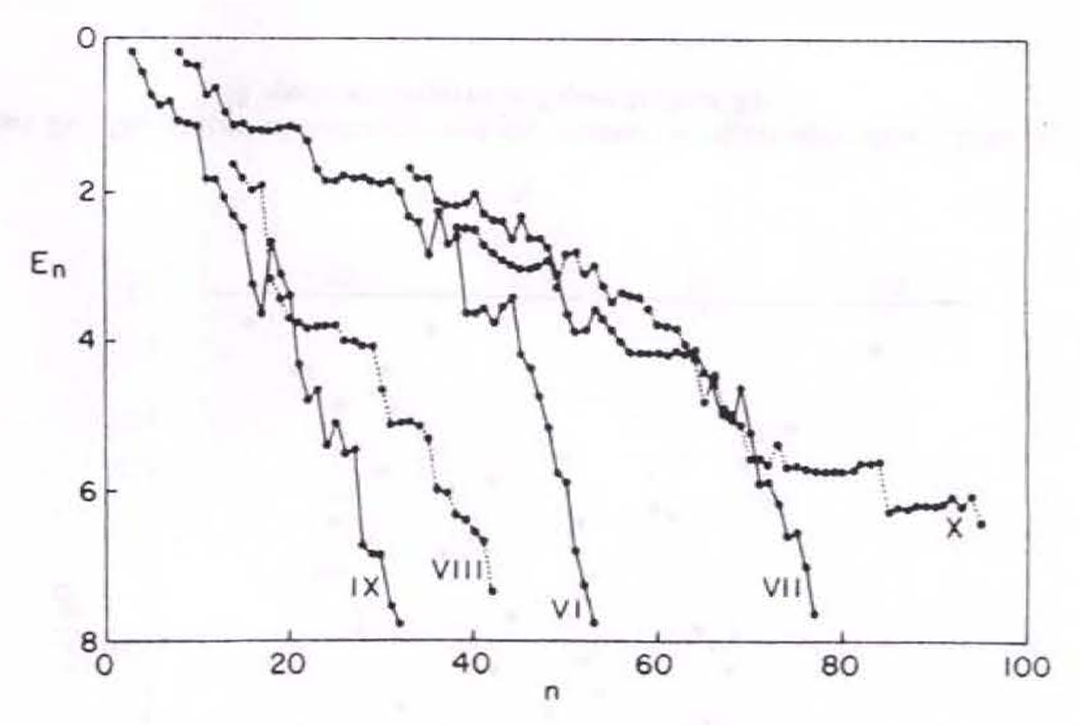
|
|
Spin Relaxation and Motional Dynamics
D.J. Schneider and J.H. Freed.
In Lasers, Molecules, and Methods, J.O. Hirschfelder, R.E. Wyatt, and R.D. Coalson, Eds.
Adv. Chem. Phys. 73, pp. 387-528 (1989).
<doi: 10.1002/9780470141229.ch10>
Publication #153
|

|
|
INTRODUCTION: The study of molecular dynamics in liquids is an active and exciting area in theoretical, computational, and experimental chemical physics. The majority of experimental techniques for studying molecular dynamics in isotropic liquids and liquid crystalline phases involve measuring the response of the system to an externally applied time-dependent perturbation, usually electromagnetic radiation. Examples of such techniques are:
magnetic resonance (electron and nuclear spin resonance),
far-infrared and infrared absorption,
dielectric relaxation,
light scattering (Raman, Rayleigh-Brillouin, etc.),
fluorescence depolarization, and
inelastic neutron scattering.
The raw data from these experiments usually reflect the underlying molecular dynamics in a rather indirect fashion. In many instances it is necessary to explicitly model the response of the system to the perturbing field to extract quantitative information on the molecular motions that modulate the experimentally observed signals. The "inversion" of the spectroscopic data to expose the dynamical information can be a difficult task, especially if the perturbing field is so strong that the system cannot respond linearly to it.
|
|
|
The Effects of Spin in Gases
F. Laloë and J.H. Freed.
Sci. Amer. 256, 94-101 (1988).
<doi: 10.1038/scientificamerican0488-94>
Publication #152
|
|
|
ABSTRACT: The nucleus of an atom can have a spin, somewhat like a tiny top. How can that spin, which is isolated from the outside world, dramatically change the properties of a gas, such as its ability to conduct heat?
|
|
|
Deuterium NMR Study of the Structure and Dynamics of the Side Chains of Several Solid Polyglutamates
E. Meirovitch, E.T. Samulski, A. Leed, H.A. Scheraga, S. Rananavare, G. Némethy, and J.H. Freed.
J. Phys. Chem. 91, 4840-4851 (1987).
<doi: 10.1021/j100302a038>
Publication #151
|
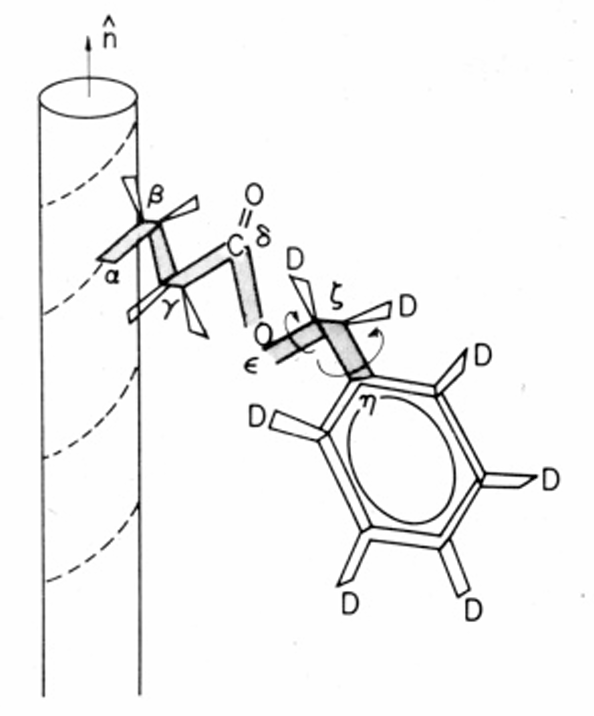
|
|
ABSTRACT: A 2H NMR study of polycrystalline powders and oriented films of several selectively deuterium-labeled poly(γ-benzyl ʟ-glutamate) (PBLG) and poly(γ-ethyl ʟ-glutamate) (PELG) homopolymers is reported. The deuterium labels on the ester side chain of PBLG indicate the presence of two polymer species below 330 K, one with benzene rings flipping rapidly about their para axis and the other with benzene rings static on the 2H NMR time scale. The relative contribution of the "mobile" population increases from about half (55%) at 240 K to nearly total (96%) at 314 K. A phase transition is detected at 330 K. Above this transition temperature, we detect an additional motion consisting of three-site jumps of the benzyl moiety among unevenly populated sites about the Oε–Cζ bond. Thus the methylene deuterons experience, on the whole, nonaxial symmetry about this bond. At 395 K this motion attains its motional-narrowing limit for all the deuterons that it affects, whereas at 340 K, only 25% of the benzyl moieties are rapidly reorienting, with 75% reorienting at a slower rate. With powders of PBLG labeled selectively at the γ-carbon position, onset of motion at the phase transition is observed. The spectral line shape indicates that, above 330 K, the γ deuteriums experience moderate (at 340 K) to rapid (at 395 K) jumps about the Cβ–Cγ bond among three unequally populated sites. The phase transition observed at 330 K, associated with the side-chain melting phenomenon, is likely to be accompanied by rotational or translational diffusion of the helices while they maintain their axial orientation. Furthermore, a comparison with our conformational analysis suggests a high degree of cooperativity between the adjacent helices. At the phase transition, these nearest-neighbor interactions would be reduced, and the NMR results are consistent with computed 3-fold rotational potentials for side-chain motion around the Cβ–Cγ and Oε–Cζ bonds, predicted from the conformational analysis of the side chains attached to the α-helix. With powders and films of PELG, the dominant motion at low temperatures (besides rapid methyl rotation) can be interpreted either as a discrete jump rotation with 3-fold symmetry but about a diffusion axis that does not coincide with the Oε–Cζ bond, or in terms of a three-site jump motion about this bond among unevenly populated sites. Motional averaging becomes very effective and isotropic in nature at about room temperature. This abrupt change from anisotropic to isotropic motion is indicative of a phase transition. Results obtained with oriented films indicate that the PBLG benzene rings orient preferentially parallel to the axes of the polymer helices, consistent with theoretical predictions, whereas the PELG ethyl groups are distributed randomly in space.
|
|
|
DSC Characterization of Nematic-Smectic and Smectic-Smectic Phase Transitions in N(p-n-Alkoxy Benzilidene)-p-n-Alkylanilines (n 0.m)
V.G.K.M. Pisipati, S.B. Rananavare, and J.H. Freed.
Mol. Cryst. Liq. Cryst. Lett. 4, 181-187 (1987).
Publication #150
|
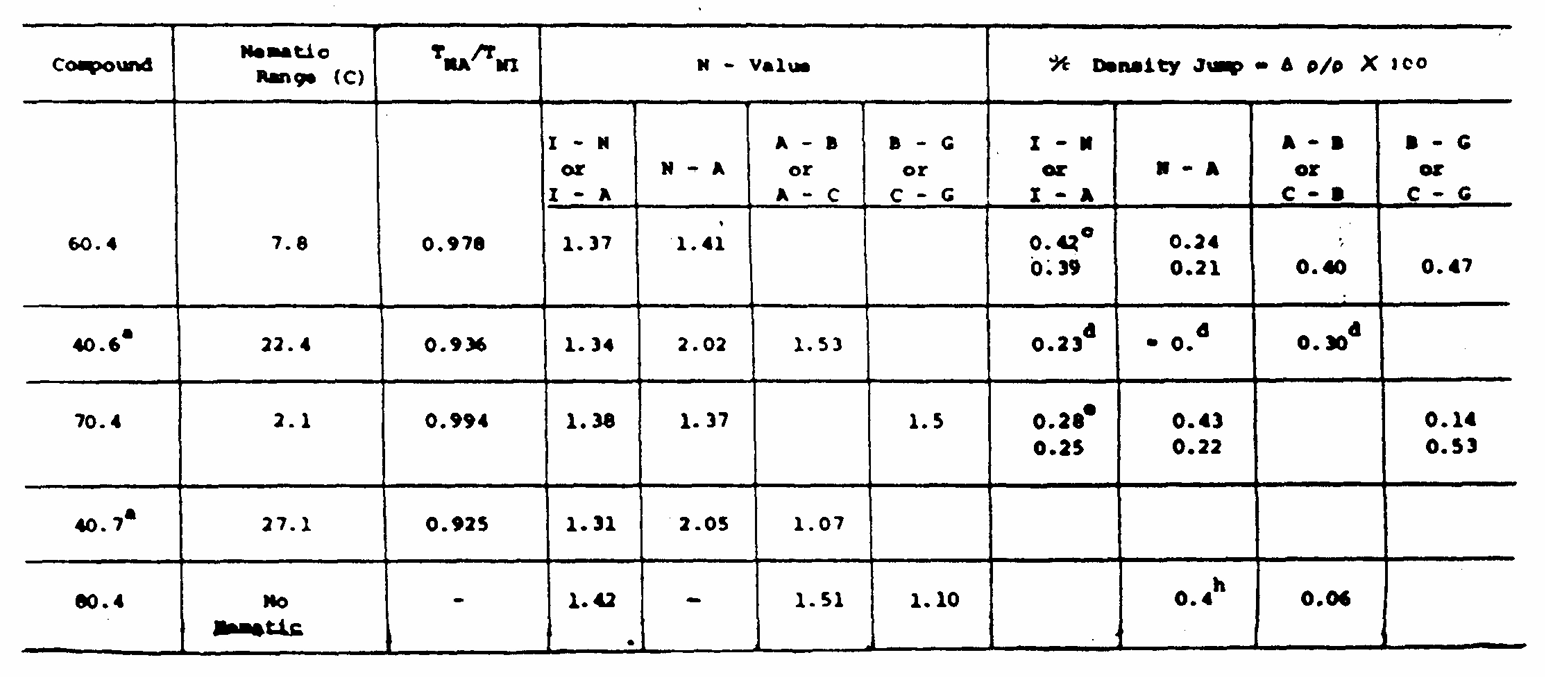
|
|
ABSTRACT: The DSC method of Navard and Haudin is applied to characterize the liquid-crystalline phase transitions in a series of n O.m compounds, which exhibit rich but subtle polymorphism. This method involves the measurement of the ratio N - h1∕h (where h1 and h are the heights of DSC transition peaks at two heating rates, one being twice the other). The particular relevance to distinguishing the order of the nematic-smectic A (NA) phase transition is emphasized.
|
|
|
Dynamic Effects of Pair Correlation Functions on Spin Relaxation by Translational Diffusion in Two Dimensional Fluids
J.P. Korb, M. Ahadi, G.P. Zientara and J.H. Freed.
J. Chem. Phys. 86, 1125-1130 (1987).
<doi: 10.1063/1.452254>
Publication #149
|
|
|
ABSTRACT: The dynamic effects of pair correlation functions (pcf) on spin relaxation by translational diffusion in infinite two-dimensional fluids are considered explicitly through a Smoluchowski equation, for the usual conditional probability, with appropriate boundary conditions, especially at the contact separation of the interacting pair of molecules. The solution of this equation by finite difference techniques permits the calculation of time correlation functions, spectral densities, and spin-relaxation rates associated with a dipolar relaxation mechanism between the spin-bearing molecules. Comparison of the two-dimensional spin-relaxation results obtained with different pcf is presented. The spectral densities and spin-relaxation rates are indeed found to be significantly altered by the pcf. For example, for a nonuniform pcf, the two-dimensional spectral densities, at (and above) the Larmor frequency ω0, are greater for translational correlation times τt that are an order of magnitude faster than τt=ω−10 which provides the maximum spectral density in bulk theory. This fast motion result is consistent with the two-dimensional dynamical results found by other techniques. Moreover for a nonuniform pcf and a single translational correlation time, two well-defined minima are found in the variation of the spin-lattice relaxation time with the diffusion coefficient. This could be very useful for interpreting the spin-relaxation data of diffusing complexes in clays, intercalation compounds, and bilayers.
|
|
|
Comments on the Interpretation of Dynamic Deuterium NMR Spectra from Solid Inclusion Compounds
E. Meirovitch, S.B. Rananavare, and J.H. Freed.
J. Phys. Chem. 91, 5014-5020 (1987).
<doi: 10.1021/j100303a026>
Publication #148
|
|
|
ABSTRACT: Guest molecules trapped within the regular voids of crystalline adducts, called inclusion compounds, are likely to experience mobility. Recent solid-state 2H NMR studies indicated these motions to be primarily discrete jumps related to the molecular symmetry, to that of the environment, or to some other geometric feature of the system investigated. In this work we offer a more consistent picture based primarily on the symmetry of the voids, but also including the molecular symmetry. We show that the 2H NMR spectra from a whole class of guest molecules can be interpreted in terms of planar threefold jumps between unequally populated sites, such that the populations follow a simple Boltzmann law. The enthalpy difference between these sites is of the order of 1-2 kcal reflecting a rather weak but nonuniform guest-host interaction.
|
|
|
Molecular Rotational Dynamics in Isotropic and Ordered Fluids Studied by ESR
Jack H. Freed
In Rotational Dynamics of Small and Macromolecules in Liquids, T. Dörfmuller and R. Pecora, Eds.
Lecture Notes in Physics, Springer-Verlag, 1987; 293, 89-142.
Publication #147
|
|
|
INTRODUCTION: ESR (like NMR) has for many years been utilized successfully in the study of rotational dynamics in liquids. Our aim in this chapter is to present a reasonably up-to-date prospectus on the subject. I have chosen to do this with a variety of recent
examples, largely from our laboratory, since I am most familiar with them.
Thus, in Section II, I describe the results of pressure and temperature-dependent spin-relaxation studies which led us to propose a new "quasi-hydrodynamic" model of rotational reorientation: the expanded-volume model. In Section III, ESR observations of deviations from Debye spectral densities at higher frequencies are described, and interpretations in terms of localized dynamic cooperativity are suggested. These studies are based upon the traditional measurements of T2's or linewidths of motionally-narrowd ESR spectra. In Section IV we introduce experiments that require a more elaborate analysis than those required for the T1 and T2 of motionally-narrowed spectra. These are slow-motional ESR studies of rotational dynamics. They are representative of a subject I like to refer to as "Beyond T1 and T2", for which considerably more sophisticated analyses may be required, but more microscopic details about the motions may be forthcoming. After first illustrating the special features of rotational motions in liquid crystals in Section V, I review in Section VI our theoretical approach for analyzing slow (and fast) motional spectra in both isotropic and ordered fluids. In the first part I summarize the formulation of the theory in terms of the stochastic-Liouville equation; then I describe the powerful modern computational algorithm that we employ to solve the lineshape problem; this is followed by some of our current ideas on modeling of rotational dynamics and dynamic cooperativity.
|
|
|
New Time Domain ESR Methods for the Study of Slow Motions on Surfaces
G.L. Millhauser, J. Gorcester, and J.H. Freed.
In Electron Magnetic Resonance of the Solid State, J.A. Weil, Ed.
Can. Chem. Soc.: Ottowa, 1987; pp. 571-598.
Publication #146
|
|
|
ABSTRACT: Cw-EPR studies of slow motions of paramagnetic species on surfaces are likely to lead to ambiguous results because there are only residual motional effects in the spectrum but significant inhomogeneous broadening due to site variations. Recently developed time-domain experiments which substantially overcome such difficulties are reviewed. In the first, one obtains a graph of the variation of T2 across the spectrum, from which the model for reorientation of spin species can be inferred. In another example, one obtains the variation of the magnetization-transfer rate TA−1 across the spectrum, which leads in the case of physisorbed N02 to a dramatic indication of anisotropic rotation. These are magnetic-field-swept two-dimensional experiments, but the very recent development of two-dimensional Fourier-Transform techniques is reviewed, since they are expected to be very useful in studies of slow motions. Also, a recent time-domain study of physisorbed H and D atoms is reviewed. It is suggested that the anomalously short temperature independent and concentration-independent phase memory time is due to surface diffusion by a tunneling mechanism.
|
|
|
Calculation of ESR Spectra and Related Fokker–Planck Forms by the Use of the Lanczos Algorithm. II. Criteria for Truncation of Basis Sets and Recursive Steps Utilizing Conjugate Gradients
K.V. Vasavada, D.J. Schneider, and J.H. Freed.
J. Chem. Phys. 86, 647-661 (1987).
<doi: 10.1063/1.452319>
Publication #145
|
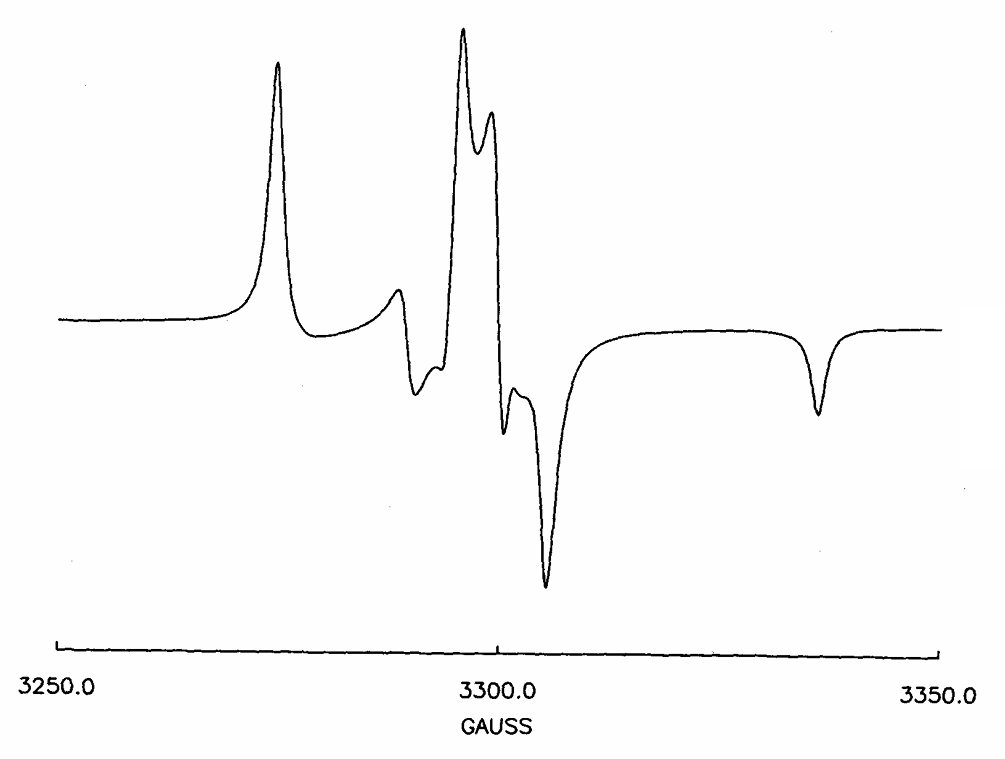
|
|
ABSTRACT: The complex symmetric Lanczos algorithm (LA) has proven to be a very efficient means of calculating magnetic resonance line shapes and spectral densities associated with Fokker–Planck forms. However, the relative importance of the various components of the basis set in an accurate representation of the spectrum and the proper number of recursive steps are not easily assessed in practice using the Lanczos algorithm. A systematic and objective procedure for the determination of optimal basis sets and number of recursive steps is developed using a generalization of the conjugate gradient method (CGM) appropriate for the type of complex symmetric matrices occuring in these problems. The relative importance of the individual basis vectors is determined by using the CGM to obtain the "solution vector" from the set of algebraic equations defining the spectrum. This is done at several values of the sweep variable (e.g., the frequency or the magnetic field). The maximum (over these values of sweep variable) for each component of the solution vector is taken to be a measure of the overall importance of the corresponding basis vector in the complete spectrum. Using this method signficant basis set truncation is conveniently possible. The number of recursive steps needed for an accurate representation of the spectrum is easily obtained by monitoring the residual in the approximate solution vector at the center of the spectrum and by recognizing the close relationship between the LA and the CGM. It is this relationship that enables construction of the Lanczos tridiagonal matrix with the CGM which can either be used to calculate the cw ESR spectrum directly or else the eigenvalues. The information obtained from the CGM can be used to "turbocharge" the LA by taking advantage of the nearly optimal basis set and number of recursive steps. Significant savings in computation time are possible, and relative savings are greatest for the most difficult problems. This is illustrated with a variety of examples of slow-motional cw ESR spectra and of the new two-dimensional electron-spin-echo technique. In keeping with the greater sensitivity of the latter technique to motional dynamics, it is consistently found to require significantly larger optimal basis sets and number of recursive steps for an accurate representation. One of the most challenging problems for both types of spectroscopy is the case of macroscopically oriented samples where the macroscopic director is tilted at an angle relative to the applied static magnetic field, since this removes much of the symmetry in the problem. This case is found to yield to very significant truncation of basis sets, and a new symmetry-based decoupling of certain basis vectors was found in this study for the particular example of a 90° tilt angle.
|
|
|
Two-Dimensional Fourier Transform ESR Spectroscopy
J. Gorcester and J.H. Freed.
J. Chem. Phys. 85, 5375-5377 (1986).
<doi: 10.1063/1.451158>
Publication #144
|
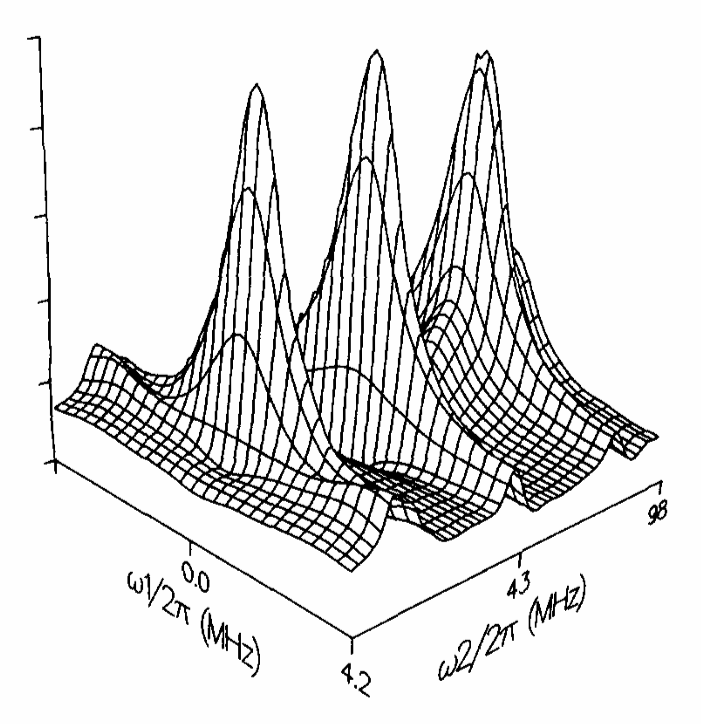
|
|
ABSTRACT: New Fourier transform ESR techniques, which permit rapid data acquisition of ESR spectra, now allow one to perform a variety of two-dimensional ESR experiments. Two prototype experiments on a fast motional nitroxide spectrum are described. The first is based upon detection of the Han echo and may be used for discriminating the homogeneous broadening of an inhomogeneous spectrum (and/or the echo envelope modulation). The second is based upon detection of the FID after the three-pulse Jeener–Ernst sequence, and it leads to cross peaks due to Heisenberg spin exhange among the hyperfine lines.
|
|
|
Linear Prediction and Resolution Enhancement of Complex Line Shapes in Two–Dimensional Electron–Spin–Echo Spectroscopy
G.L. Millhauser and J.H. Freed.
J. Chem. Phys. 85, 63-67 (1986).
<doi: 10.1063/1.451595>
Publication #143
|
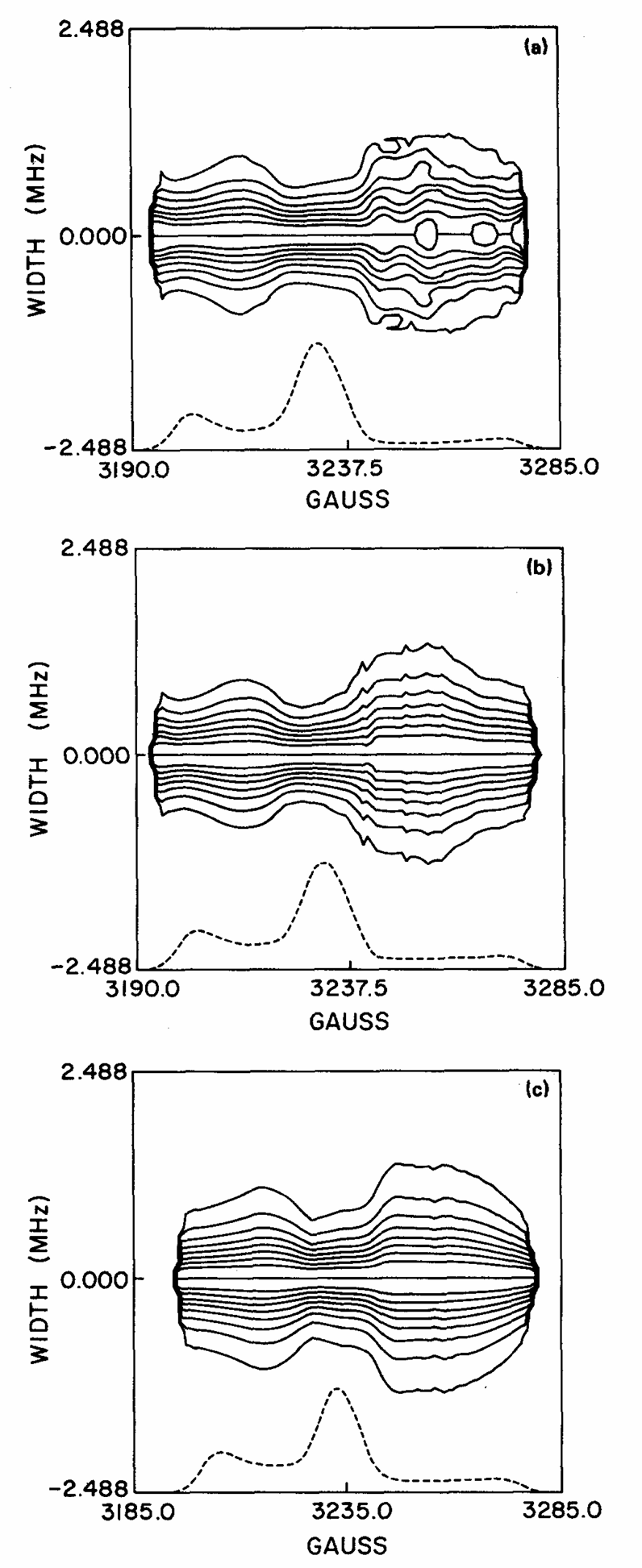
|
|
ABSTRACT: A new type of two-dimensional electron-spin-echo (2D-ESE) spectroscopy was recently shown to be useful for studying slow molecular motions in liquids. A recently developed method of spectral enhancement based upon linear prediction with singular value decomposition (LPSVD) is applied in the present work to dramatically improve the signal-to-noise ratio and to correct for finite dead time in the data from this 2D-ESE experiment. This permitted a more accurate comparison between theory and experiment. Good agreement is now obtained with a model of nearly isotropic Brownian motion for tempone in glycerol/water solvent.
|
|
|
|
|
The Lanczos Algorithm in Molecular Dynamics: Calculation of Spectral Densities
G. Moro and J.H. Freed.
In Large Scale Eigenvalue Problems, J. Cullum and R.A. Willoughby, Eds.
Mathematics Studies Series, 127, 143-161 (1986). (Elsevier Science Publishers B. V., North-Holland).
<doi: 10.1016/S0304-0208(08)72644-6>
Publication #141
|
|
|
ABSTRACT: In the past the Lanczos algorithm has proven to be a very useful tool in the study of molecular dynamics. It enables efficient calculation of the spectral densities which are essential in the interpretation of spectroscopic or scattering experiments. In the present work, the use of the Lanczos algorithm in the calculation of the spectral densities is analyzed in a general fashion. Its application to problems characterized by complex symmetric matrices, which are normally encountered in the analysis of magnetic resonance experiments, is then recovered as a particular case. After a discussion of the factors influencing the convergence in the calculation of spectral densities, the implementation of the Lanczos algorithm with non-orthogonal basis functions is considered in relation to molecular systems having hindered degrees of freedom.
|
|
|
Spectral Rotation in Pulsed ESR Spectroscopy
J.P. Hornak and J.H. Freed.
J. Magn. Reson. 67, 501-518 (1986).
<doi: 10.1016/0022-2364(86)90387-2>
Publication #140
|
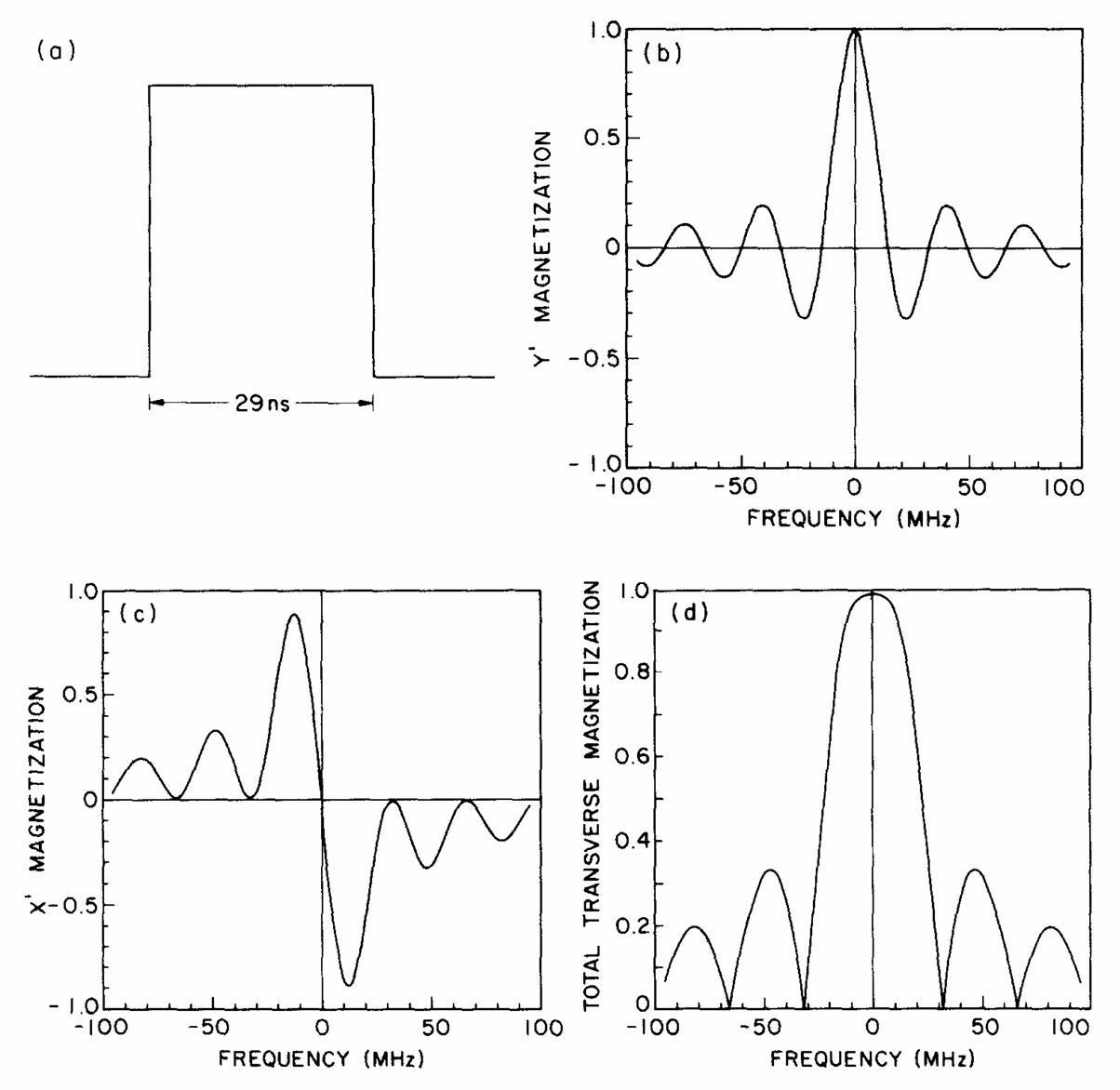
|
|
ABSTRACT: The technique of pulsed Fourier transform NMR spectroscopy is well developed and understood. The equivalent technique for ESR has been slower to be developed, but recent developments in ESR instrumentation have now made FT ESR possible. In this paper we consider the requirements for rotating the magnetization associated with an ESR spectrum by a microwave pulse. In addition, techniques for reconstructing the frequency (field)-domain spectrum from the time-domain spectra are described. These techniques are applied to obtain a FT spectrum from the electron spin-echo decay of γ-irradiated quartz and a FT spectrum from the free induction decay of the 2,5-di-t-butyl p-benzosemiquinone radical recorded in 50 and 25 ms, respectively, of real time.
|
|
|
Diffusion Coefficients in Anisotropic Fluids by ESR Imaging of Concentration Profiles
J.P. Hornak, J.K. Moscicki, D.J. Schneider, and J.H. Freed.
J. Chem. Phys. 84, 3387-3395 (1986).
<doi: 10.1063/1.450221>
Publication #139
|
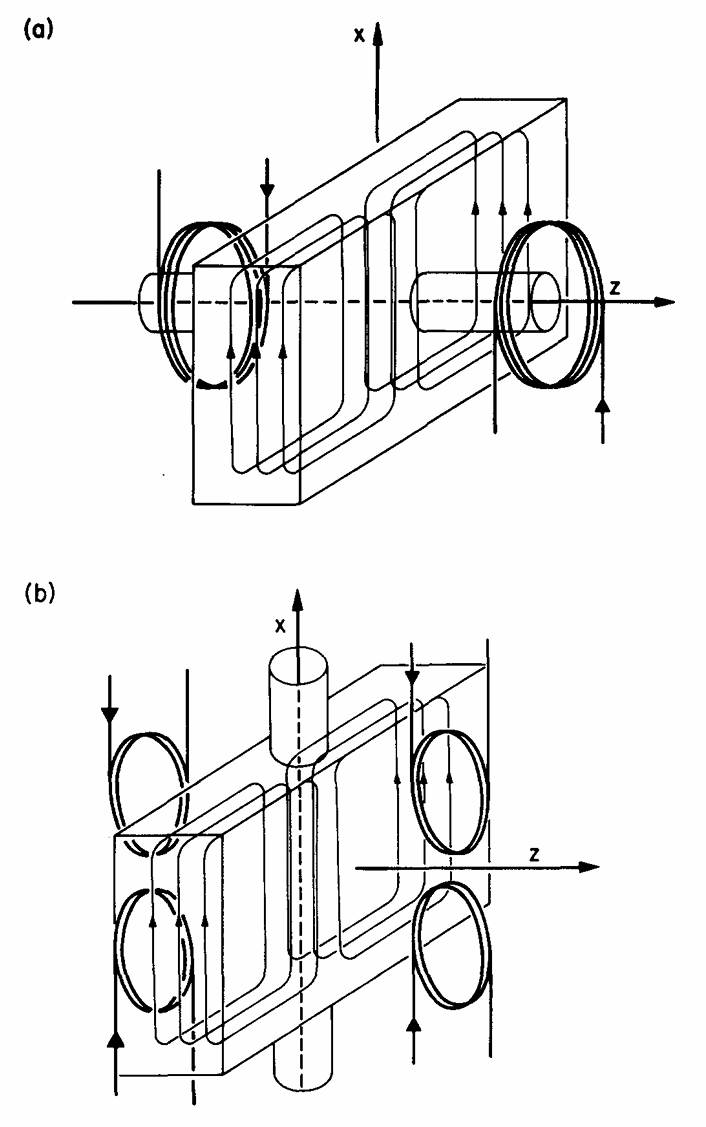
|
|
ABSTRACT: We report on the use of ESR imaging for determining the translational diffusion constants of typical ESR spin probes in ordered and isotropic solvents. A discussion is given for a Fourier deconvolution method for determining the correct concentration profile if there is more than one hyperfine line in the spectrum of the radical as well as a spatial dependence of the spectrometer sensitivity. A simple but approximate subtraction deconvolution method is also presented. The diffusion constants for 4-oxo-2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPONE) in the nematic and isotropic phases of the liquid crystal p-pentylbenzylidine-p-butylanaline (5,4) were determined. In the isotropic phase the diffusion coefficient is isotropic (D=2.5×10−6 cm2 s−1, at 50 °C), while in the nematic phase it was found to be mildly anisotropic. The diffusion coefficients for motion perpendicular and parallel to the director axis in the nematic phase are respectively, D⊥ =9.0×10−7 cm2 s−1, D∥ =6.4×10−7 cm2 s−1 at 27 °C. This is interpreted in terms of some smectic-like character in the nematic phase of 5,4. Possible improvements in the technique are also discussed.
|
|
|
Spin-Waves in Spin Polarized H↓
J.H. Freed
Ann. de Phys. 10, 901-921 (1985).
<doi: 10.1051/anphys:01985001006090100>
Publication #138
|
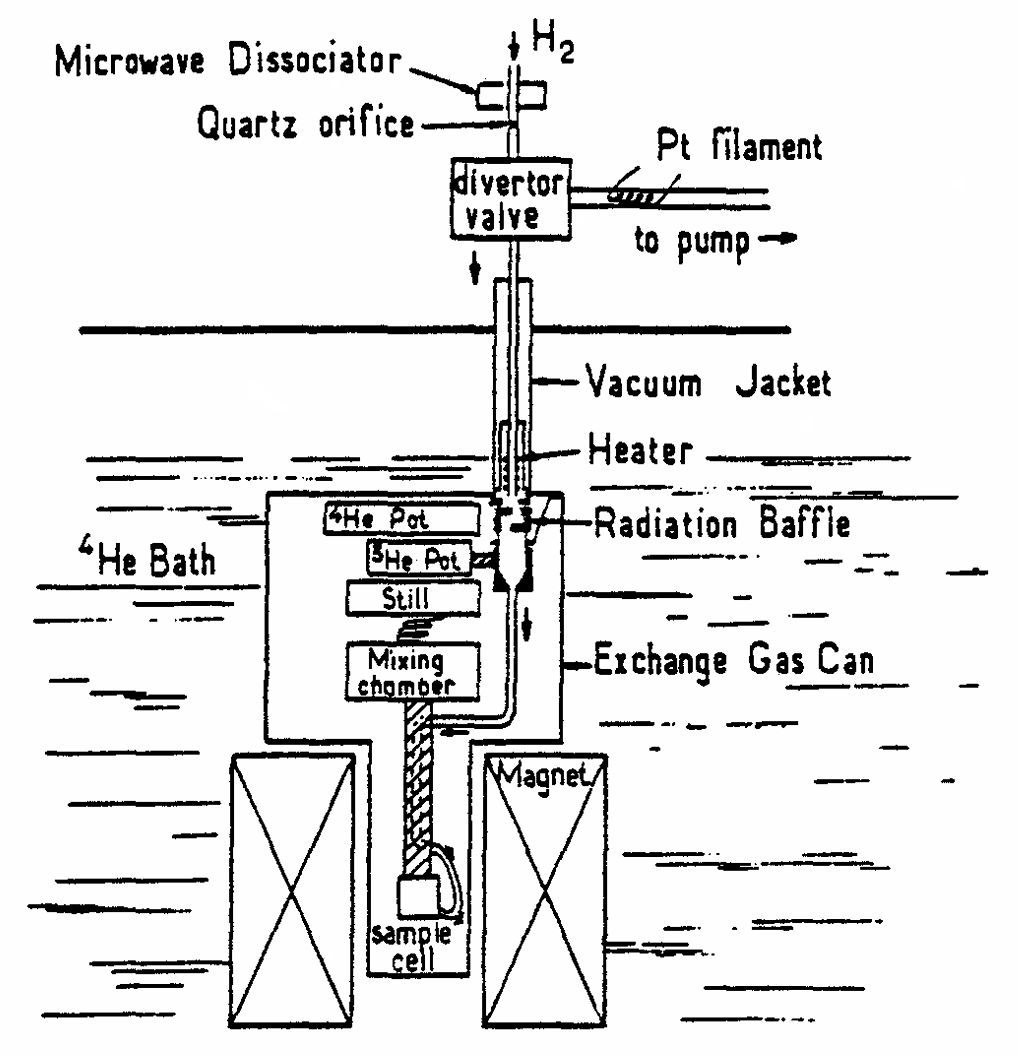
|
|
ABSTRACT: This is a short review of previous published and unpublished results of our Cornell University group pertaining to the observations of damped nuclear spin-wave modes in spin-polarized atomic hydrogen gas.
|
|
|
Surface-Suppressed Electron Resonance Spectroscopies
P.G. Barkley, J.P. Hornak, and J.H. Freed.
J. Chem. Phys. 84, 1886-1900 (1986).
<doi: 10.1063/1.450437>
Publication #137
|
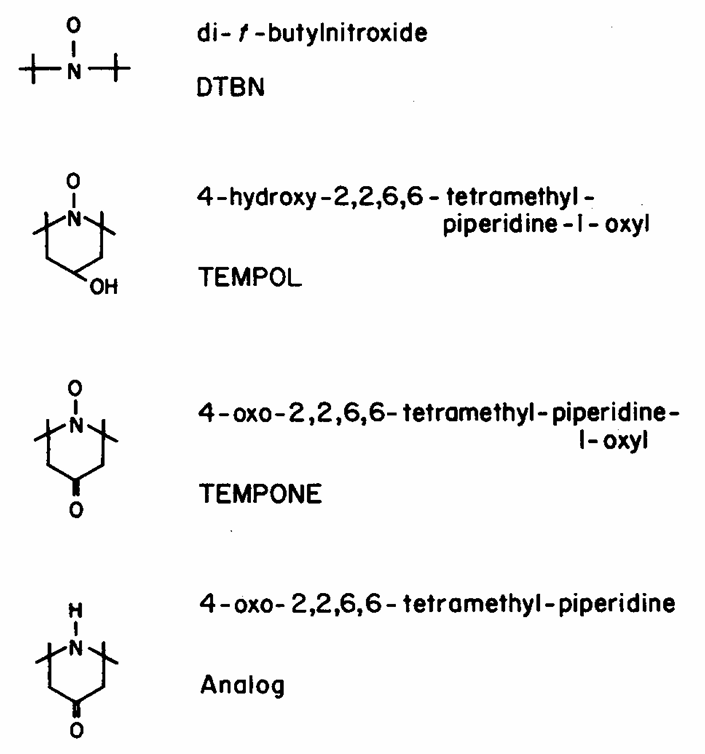
|
|
ABSTRACT: Surface-suppressed electron resonance spectroscopies (SSERS) refers to the phenomenon in which stable paramagnetic radicals adsorbed on clean (noble) metal surfaces have their ESR signal suppressed. This phenomenon is studied in some detail by ultra-high-vacuum ESR (UHV-ESR) and cyclotron resonance from microwave-induced secondary electron emissions (CREMSEE) in combination with conventional thermal desorption. The UHV-ESR is performed in situ on the inner surface of the microwave cavity after leaking in stable nitroxides as previously described by Nilges and Freed. In the SSERS phenomenon, the first layers of nitroxide deposited on the metal surface do not give rise to an observable ESR signal, even though a significant decrease in the CREMSEE microwave power threshold, Pt, is observed. It is consistently found (for several nitroxides and noble metals) that an ESR signal is observed only when Pt has dropped to of the order of 20 (±10)% of its initial value. The ESR signal then increases monotonically with increased dosage. Also, a correlation between these measured reductions in Pt, and the estimated surface coverages required for initial observation of the ESR signal, is suggested by our results. It may be that SSERS is (partly) due to the first layer of nitroxide interacting strongly by exchange forces to the surface conduction band of the metal, so that the ESR signal is too broadened to be observed. Subsequent nitroxide layers may also be affected through their interaction with previous layers by weaker exchange forces. This is consistent with experiments in which a related but diamagnetic species is utilized to form the initial adsorbed layers, and it is found to act as an "insulator" for the subsequent nitroxide layers. (On the other hand, surfaces pretreated with 02 or 02∕H2O mixtures had very little effect on SSERS observed with Ag surfaces, although it had some effect, in the sense of a weak insulator, with Cu surfaces.) The change in ESR signal upon warming was correlated with the observed pressure changes. In some cases there are unusual nonmonotonic variations of the ESR signal strength, inconsistent with observed desorption of nitroxide, that are also believed to be due to SSERS. The possible role of temperature-dependent surface wetting effects is briefly considered.
|
|
|
Electron Spin Echoes with a Loop-Gap Resonator
J.P. Hornak and J.H. Freed.
J. Magn. Reson. 62, 311-313 (1985).
<doi: 10.1016/0022-2364(85)90065-4>
Publication #136
|
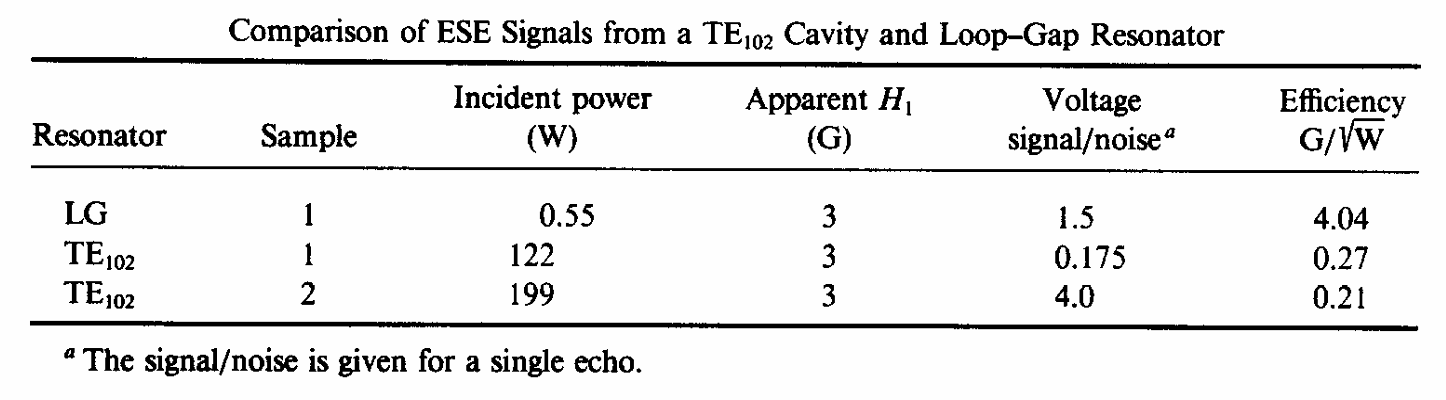
|
|
INTRODUCTION: We report our preliminary findings on the use of a split-ring or loop-gap resonator in electron spin echo (ESE) spectroscopy at 9 GHz. The loop-gap (LG) resonator of Froncisz and Hyde was reported to have several features which could make it more advantageous than the overcoupled TE102 microwave cavity traditionally used in ESE experiments. Among these are the large uniform microwave magnetic field H1 per incident microwave power, broadband (low Q), small sample volume, high filling factor, and high field homogeneity over the sample. These advantages could make it possible with short pulse lengths to rotate an entire nitroxide spin probe spectrum (30 G) due to the large H1. In addition, conventional ESE experiments could be carried out with a 1 W klystron rather than a traveling wave tube amplifier (TWTA), thus significantly reducing the cost of an ESE spectrometer. Here we compare our findings on the LG resonator to an overcoupled TE102 cavity typical of that used in ESE spectroscopy.
A two-slit loop-gap resonator constructed out of brass and plated with silver was used rather than Macor (Corning Glass) since field modulation is not required in the ESE experiments, and it has superior tolerance to temperature extremes. The dimensions of the resonator were 6.6 mm o.d., 1.2 i.d., 0.1 mm slit width, and 4.5 mm in length. The slits were filled with Teflon, resulting in resonance at 8.85 GHz. The resonator was inductively coupled to the microwave bridge by a loop through the 12.7 mm i.d. outer shield around the resonator. The loaded Q of the LG resonator at critical coupling was approximately 400. The TE102 cavity was a Varian V-4532 with a Gordon coupler and 1.0 cm × 0.72 cm × 0.127 mm thick rectangular coupling hole of silver-plated brass. This configuration allowed overcoupling and gave the cavity a loaded Q of approximately 400. Both the LG resonator and TE102 cavity were overcoupled such that the voltage reflection coefficient was approximately 0.85 (a VSWR of 12) so the calculated overcoupled Q was 65. The details of the ESE spectrometer are described elsewhere.
|
|
|
Spin Waves in Spin-Polarized Hydrogen Gas
D.M. Lee and J.H. Freed.
Physics Today 38, S-18 (1985).
Publication #135
|
|
|
INTRODUCTION: Spin waves are quantum phenomena most commonly found in condensed matter, where the overlap (in terms of quantum mechanics) of adjacent atoms leads to strong correlations between neighboring spins. A spin wave corresponds to a wavelike propagation through a medium of a local tilt of spins from some preferred orientation. Previously spin waves had been observed in a wide variety of systems, including ferromagnetic materials, antiferromagnetic materials, and even in liquid 3He at very low temperatures.
Surprisingly, an experiment recently performed at Cornell University has revealed that it is possible to excite spin waves at temperatures of about 0.4 K. in a highly rarefied gas of spin-polarized hydrogen (1016 atoms per cm3, corresponding to about one-thousandth the density of air at sea level). In such a dilute gas an atom spends most of its time far away from its neighbors, so that the quantum overlap found in condensed systems occurs only on the very rare occasions when two atoms in the gas collide. It was shown theoretically by physicists in France and the Soviet Union that under special conditions, even the occurrence of these rare collisions in a dilute gas could lead to spin waves. In the case of atomic hydrogen the most important criterion is that two colliding atoms be essentially indistinguishable. Since hydrogen atoms are each composed of one electron and one proton, the indistinguishability requirement is met by arranging for all the electron spins and all the proton spins in a small region of the sample to be lined up in the same direction along an axis usually taken as the z axis.
|
|
|
Electron Spin Resonance and Electron-Spin-Echo Study of Oriented Multilayers of Lα-Dipalmitoylphosphatidylcholine Water Systems
L. Kar, E. Ney-Igner, and J.H. Freed.
Biophys. J. 48, 569-595 (1985).
<doi: 10.1016/S0006-3495(85)83814-5>
PMID:
2996648
PMCID:
PMC1329335
Publication #134
|
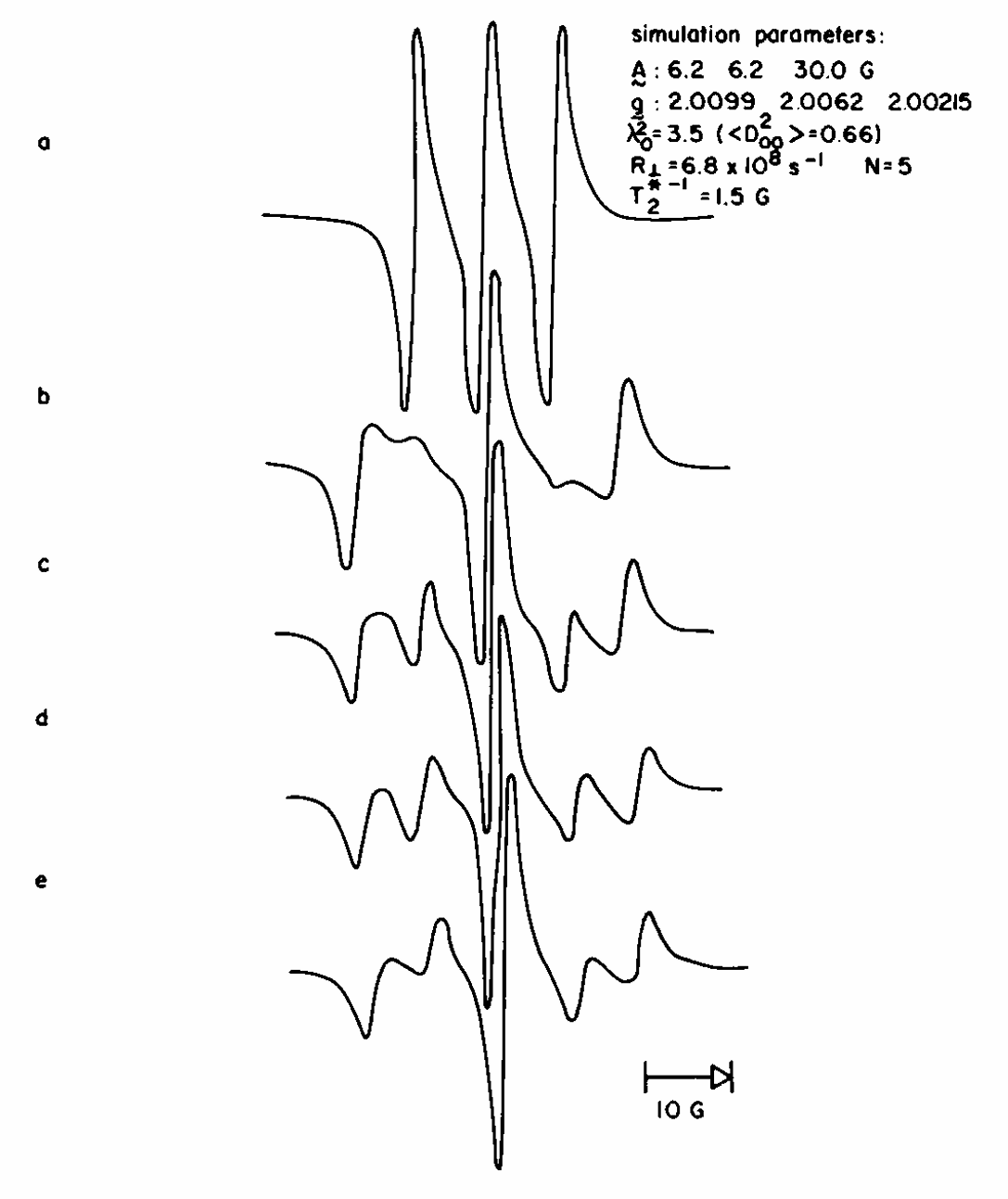
|
|
ABSTRACT: A detailed electron spin resonance (ESR) study of spin-labeled-oriented multilayers of Lα-dipalmitoylphosphatidylcholine (DPPC) water systems for low water content (2-10% by weight) is reported with the purpose of characterizing the dynamical and structural properties of model membrane systems. Emphasis is placed on the value of combining such experiments with detailed simulations based on current slow-motional theories. Information is obtained regarding ordering and anisotropic rotational diffusion rates via ESR lineshape analysis over the entire motional range, from the fast motional region through the moderately slow and slow to the rigid limit. This includes the low-temperature gel phase, the liquid crystalline Lα(1) phase and what appears to be a third high-temperature phase above the Lα phase. Cholestane (CSL) and spin-labeled DPPC (5-PC, 8-PC, and 16-PC) have been used to probe different depths of the bilayer. While CSL and 5-PC both reflect the high ordering of the bilayer close to the lipid-water interface, CSL appears to be located close enough to the water for the nitroxide to be involved in hydrogen bonding with water molecules. 16-PC reflects the relatively low ordering near the tail of the hydrocarbon chain in the bilayer. Quantitative estimates of ordering and motion are obtained for these cases. The results from CSL indicate that close to the lipid-water interface the DPPC molecule is oriented approximately perpendicular to the bilayer in these low water-content systems. However, all three labeled lipid probes indicate that the hydrocarbon chain of DPPC may be bent away from the bilayer normal by as much as 30° and this evidence is stronger at low temperatures. When cholesterol is added to the DPPC-water system at a concentration ≥ 2.5 mol %, the ordering is greatly increased although the rotational diffusion rate remains almost unaffected in the gel phase. Electron spin echoes (ESE) are observed for the first time from oriented lipid-water multilayers. Results obtained from cw ESR lineshape analysis are correlated with data from ESE experiments, which give a more direct measurement of relaxation times. These results indicate that for detection of very slow motions (close to the rigid limit) ESE experiments are more sensitive to dynamics than continuous wave ESR for which inhomogeneous broadening becomes a major problem.
|
|
|
Electron Spin Resonance Studies of Lipid-Gramicidin Interactions Utilizing Oriented Multibilayers
H. Tanaka and J.H. Freed.
J. Phys. Chem. 89, 350-360 (1985).
<doi: 10.1021/j100248a034>
Publication #133
|
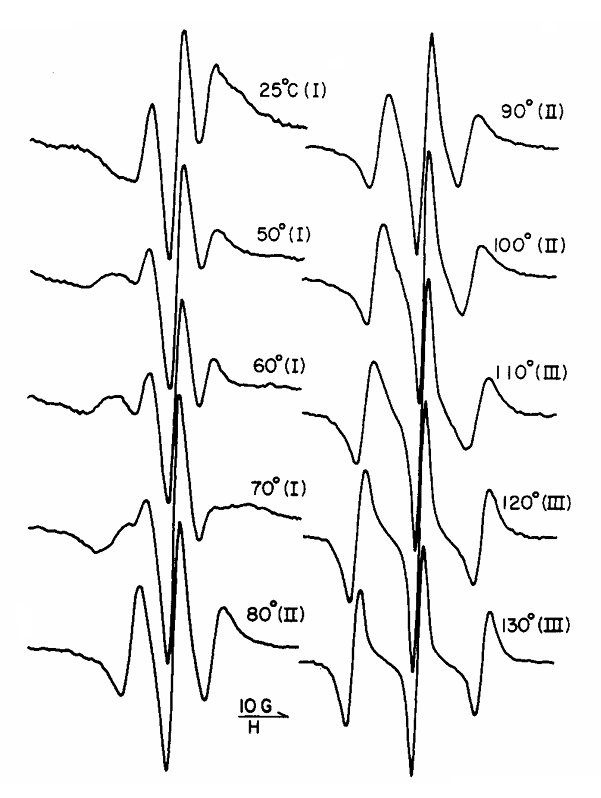
|
|
ABSTRACT: Macroscopically well-aligned lipid (DPPC) samples of ≤10 wt % water, which contain 0-4 mol % of the polypeptide gramicidin A (GA) and which yield reproducible ESR spectra from spin-labeled lipids and from cholestane probe, have been prepared by a recently developed method. The effect of GA was analyzed in detail by first subjecting the extensive ESR spectra obtained to the methods of spectral analysis previously developed by Freed and co-workers. The great value of combining studies on oriented samples with such quantitative theoretical analysis is emphasized. In the gel phase a heterogeneity was found, which could be analyzed as two types of regions: one of the bulk lipids and one of boundary or disordered lipids. The principal difference between the two is the greatly reduced ordering of the latter, but with little difference in fluidity. For low concentration of GA (and 7 wt % water), it is estimated that there are 60-80 disordered lipids per GA dimer, or a disordered region in the bilayer extending radially about three lipid molecules. The disordered region is reduced by increased weight percent water but does not appear to be affected by temperature. In the liquid crystalline phase the heterogeneity is no longer evident in the ESR spectra. Instead the ordering of all the lipids is significantly reduced by the addition of GA, but motional rates are only slightly reduced. In the high-temperature, low-ordering phase previously studied, addition of GA actually increases the ordering. It is concluded from these results that the principal lipid-GA interaction is that of a boundary effect such that the GA induces disorder in low-temperature and low-water-content lipids but it induces order for high-temperature and high-water-content (Le., less ordered) lipids. It has only slight effects on lipid fluidity, in general reducing only slightly the rates of rotational reorientation. No spectral features usually attributed to "immobilized" lipids were observed in the spectra from the well-aligned samples even when they were present in spectra from dispersions of the same material content at the highest GA concentrations. If these spectral features are interpreted as associated with "trapped lipids" due to aggregation of polypeptide, then it follows that the well-aligned samples do not permit such aggregation. These results also clearly show that the heterogeneity induced by GA at very low concentrations is a distinctly different phenomenon from the one usually assigned to "immobilized" or "trapped" lipids.
|
|
|
Electron Spin Resonance Studies on Ordering and Rotational Diffusion in Oriented Phosphatidylcholine Multilayers: Evidence for a New Chain-Ordering Transition
H. Tanaka and J.H. Freed.
J. Phys. Chem. 88, 6633-6644 (1984).
<doi: 10.1021/j150670a027>
Publication #132
|
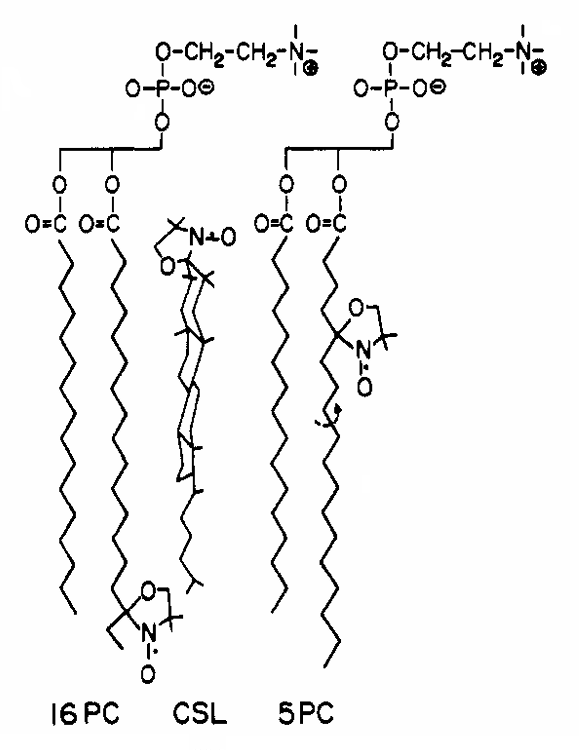
|
|
ABSTRACT: An alignment technique based on compression has been developed for preparation of planar oriented lipid samples for ESR. The ESR spectra of defect-free samples prepared by this technique could be studied over a wide temperature range, showing gel and liquid-crystalline phases of DPPC and DMPC. The ESR spectra of spin-labeled DPPC's and cholestane clearly indicate a new phase transition occurring at 100-110 °C in the liquid-crystalline phases of DPPC and DMPC hydrated to 3 wt % water with somewhat higher temperatures for higher water content. The extensive spectra that were obtained could be analyzed in detail with a model of anisotropic molecular rotation in a mean orienting potential by using the methods of spectral analysis previously developed by Freed and co-workers. On the basis of the results on molecular ordering and rates of rotational diffusion, the new phase transition is characterized as one in which orientational order is significantly reduced but the rate of molecular motions is not drastically changed. This is compared with a recent theoretical model, which predicts a similar high-temperature transition. The present results also reveal some new aspects regarding hydrocarbon chain motion and disorder in the crystalline phase.
|
|
|
Spin Echoes in Spin Polarized Hydrogen
J.S. Denker, N. Bigelow, D. Thompson, J.H. Freed, and D.M. Lee.
Proc. 17th Intl. Conf. on Low Temp. Physics 549-550 (1984).
Publication #131
|
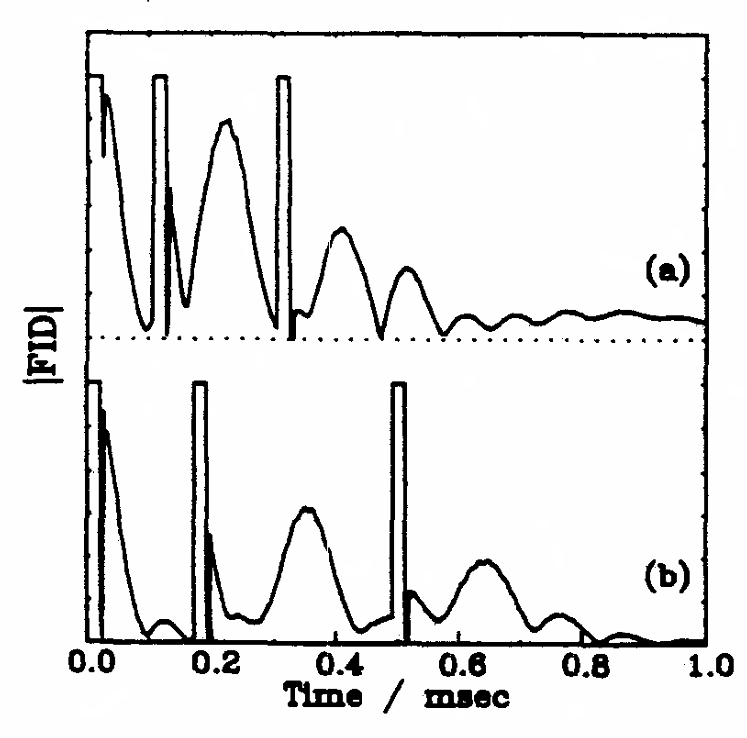
|
|
ABSTRACT: We have conducted preliminary spin echo experiments in spin polarized hydrogen. The heights of the echoes follow a remarkable pattern, and we often see a regular series of additional peaks in the FID envelope which resemble multiple echoes. We conjecture that molecular fields, with due regard for boundary conditions, could account for most of the observed effects.
|
|
|
Observation of Spin Waves in Spin Polarized Hydrogen
B.R. Johnson, N. Bigelow, J.S. Denker, L.P. Lévy, J.H. Freed, and D.M. Lee.
Proc. 17th Intl. Conf. on Low Temp. Physics 451-452 (1984).
Publication #130
|
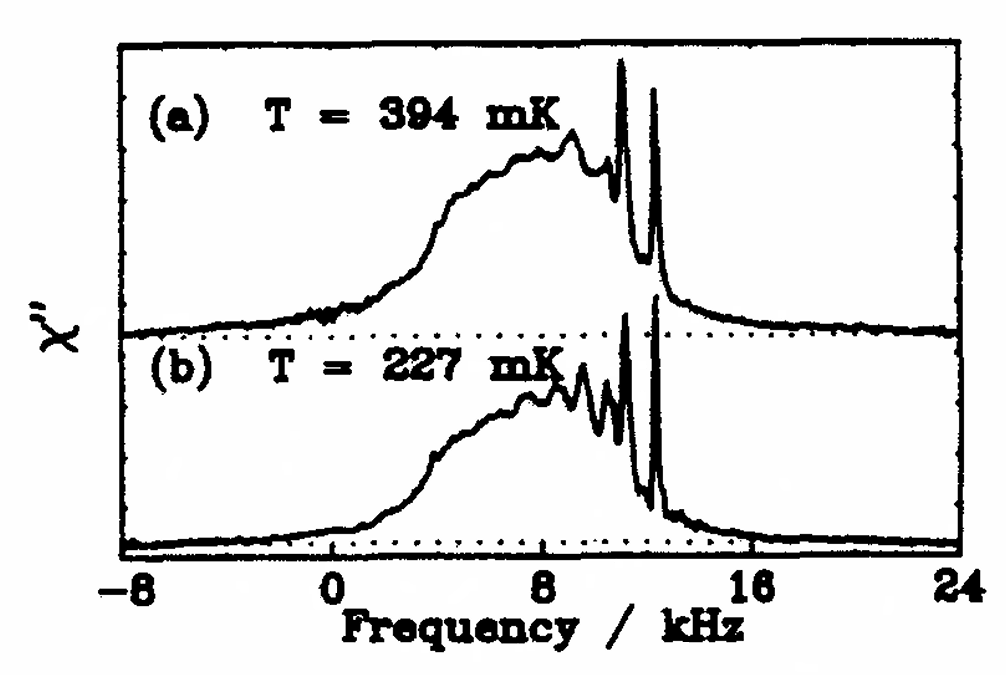
|
|
ABSTRACT: We report the results of continuing experiments on the spin transport properties of a dilute quantum gas, spin polarized hydrogen H↓. Spin wave resonances are prominent features of the pulsed Fourier transform NMR spectrum. For small tipping angles, the dependence of the spectrum on polarization and temperature are found to be in good qualitative agreement with theory. Preliminary results are presented on the large tipping angle spectrum, which exhibits a number of features not observed in the small angle spectrum.
|
|
|
Molecular Dynamics at the Nematic to Smectic A Phase Transition: An ESR Study
S.A. Zager and J.H. Freed.
Chem. Phys. Lett. 109, 270-275 (1984).
<doi: 10.1016/0009-2614(84)85733-4>
Publication #129
|
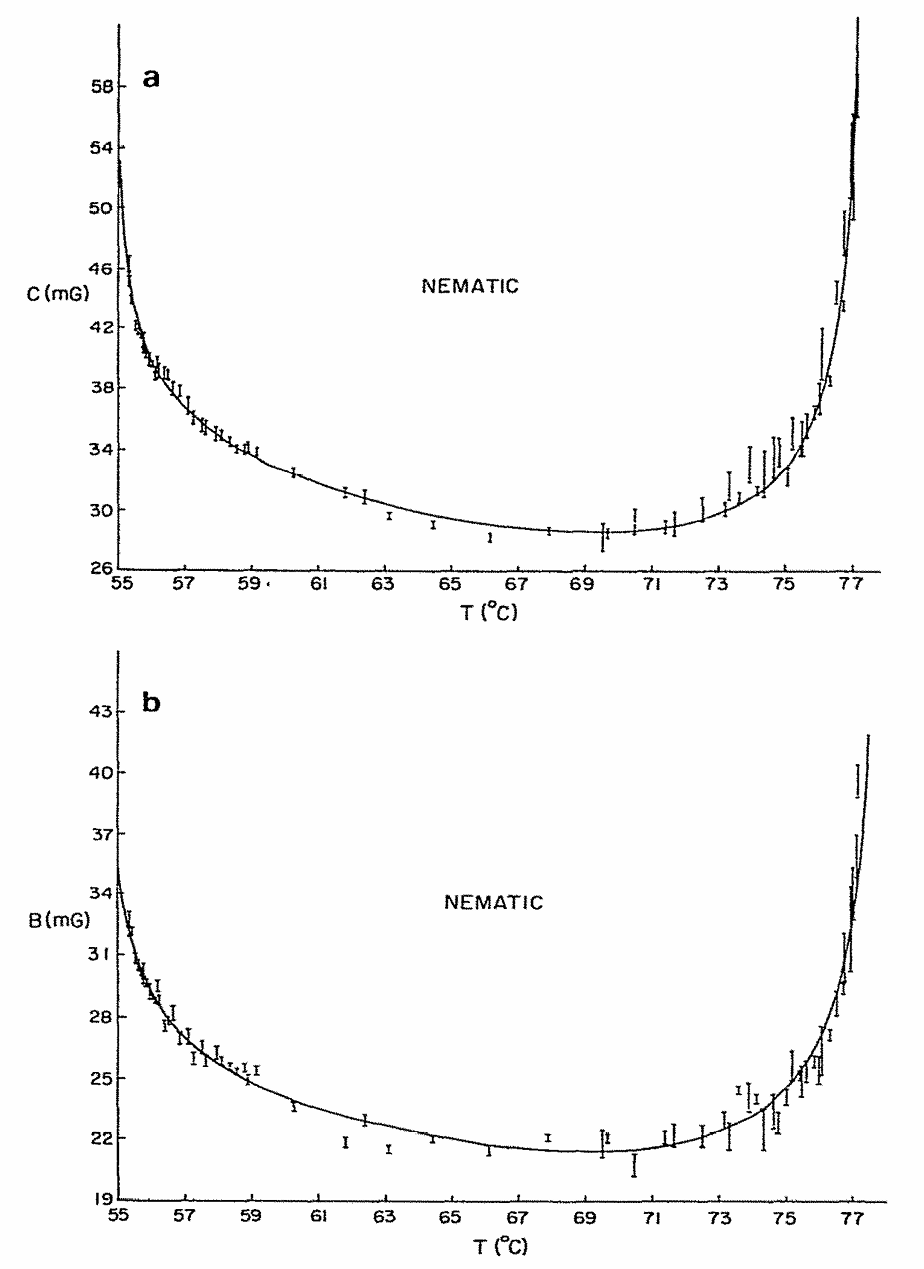
|
|
ABSTRACT: In an electron-spin-relaxation study of a spin probe dissolved in a liquid crystal, the linewidth parameters are observed to diverge in the nematic phase with critical exponent close to 1/3 as the nematic-smectic A transition is approached. This is interpreted in terms of a model wherein pre-transitional smectic fluctations modulate the spin-relaxation parameters of the probe.
|
|
|
Detection of Slow Motions in Oriented Lipid Multilayers by Two-Dimensional Electron-Spin-Echo Spectroscopy
L. Kar, G.L. Millhauser, and J.H. Freed.
J. Phys. Chem. 88, 3951-3956 (1984).
<doi: 10.1021/j150662a016>
Publication #128
|
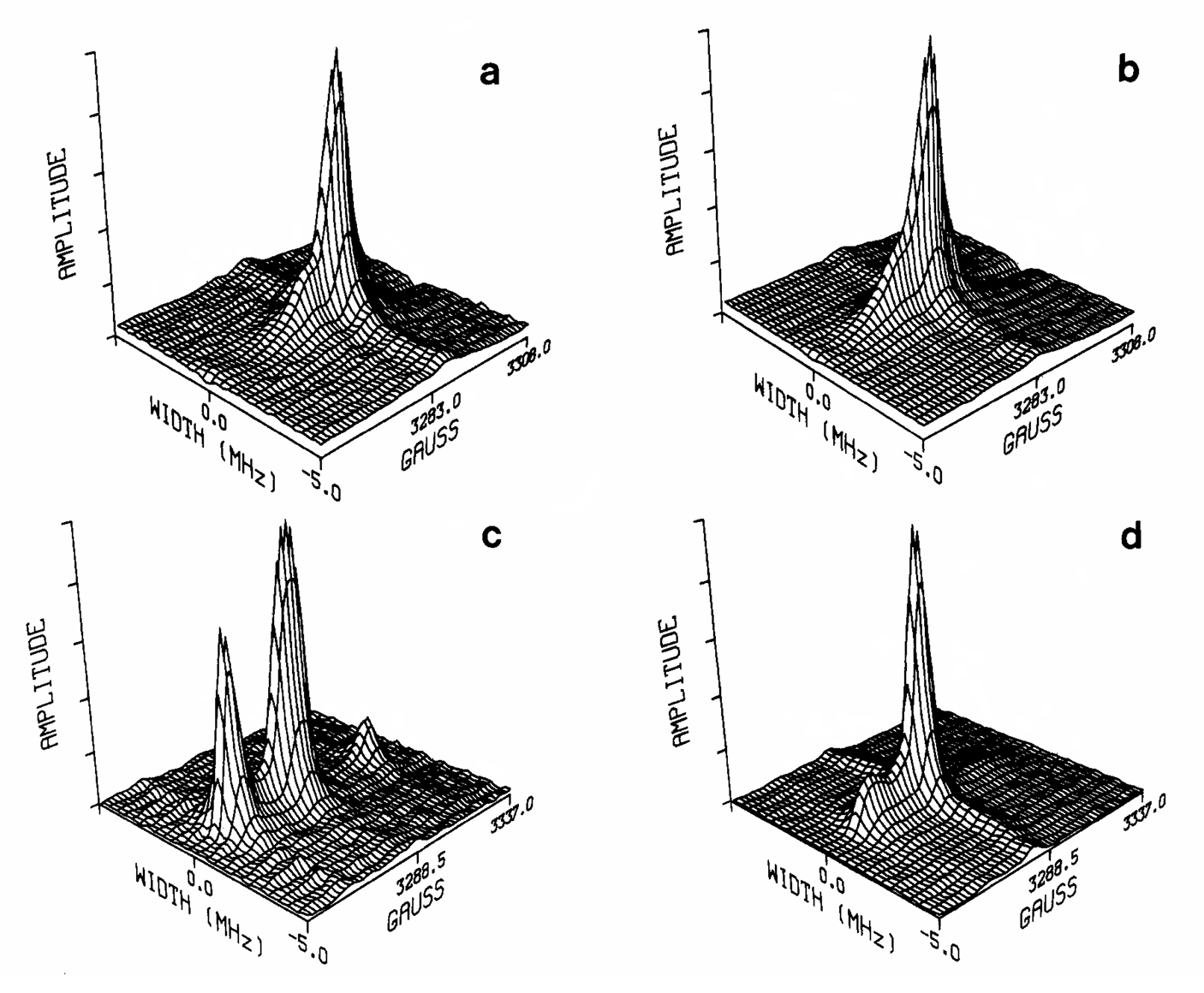
|
|
ABSTRACT: The considerable potential of two-dimensional electron-spin-echo (2D-ESE) spectroscopy in the study of structure and dynamics in oriented media for spin probes undergoing slow molecular motions is demonstrated on a model membrane system doped with spin probe. A 90-τ-180 echo sequence is used to detect a variation of the homogeneous line width (T2-1) across the "echo-induced ESR spectrum". Simulations using recently developed methods are found to be very sensitive to parameters describing the rotational diffusion and the ordering potential. Rotational diffusion rates of the order of 104 s-1 are detected at temperatures below 0 °C, and a model of Brownian motion and a high ordering is found to characterize this smectic system consisting of low water content oriented multilayers of dipalmitoyl phosphatidylcholine (DPPC) doped with cholestane (CSL) spin probe. The great sensitivity of this 2D-ESE technique to a motional region inaccessible to cw studies is emphasized.
|
|
|
Analysis of Slow-Motional Electron Spin Resonance Spectra in Smectic Phases in Terms of Molecular Configuration, Intermolecular Interactions, and Dynamics
E. Meirovitch and J.H. Freed.
J. Phys. Chem. 88, 4995-5004 (1984).
<doi: 10.1021/j150665a041>
Publication #127
|
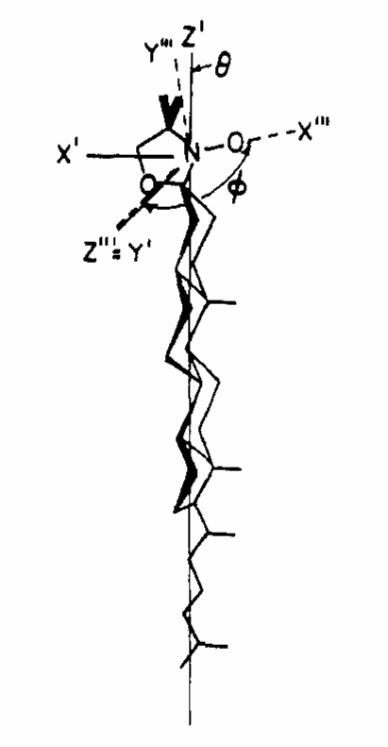
|
|
ABSTRACT: ESR spectra from two oxazolidine derivative spin probes (CSL and 1,14-stearic acid) dissolved in the smectic phase of S2 were carefully analyzed and the results interpreted in terms of ordering characteristics, molecular conformation, and dynamics by using powerful and comprehensive spectral simulation techniques. The rigid CSL is highly ordered in the smectic A phase as expected for strong interactions with the rigid aromatic cores of the liquid-crystal molecules and shows reorientational motion well approximated as Brownian in the mean potential of the solvent molecules. The principal axes of ordering of CSL are found to be tilted with respect to the principal axes of the magnetic tensor, and the Euler angles specifying this tilt could be determined, because of the sensitivity of the slow-motional spectra to these parameters. The considerable sensitivity of these spectra to the shape of the orienting potential is also demonstrated, and values for coefficients in the expansion of the potential in the spherical harmonics through L = 4 are estimated. The flexible 1,14-stearic acid probe is only weakly ordered in the smectic phase. It shows an anisotropy in its rotational diffusion tensor that is smaller than predicted for an extended all-trans conformation, suggesting an average configuration with a decreased length-to-width ratio. However, the end segment containing the N-O group appears to be in an all-trans configuration, judging by the observed collinearity between the principal symmetry axis of diffusion and ordering with that of the hyperfine tensor. Its low ordering and reduced activation energy for reorientation in the smectic phase suggest primary coupling to the alkyl chain region of the solvent molecules. The spectral simulations are improved by introducing asymmetry in the viscosity, but related anomalies suggest the likelihood of some other mechanism such as a fluctuating torque model. A comparison of the results of this and other studies is made to show how, by the use of different spin probes, one can obtain insights into the intermolecular interactions operating in anisotropic fluids.
|
|
|
Analysis of Protein-Lipid Interactions Based on Model Simulations of Electron Spin Resonance Spectra
E. Meirovitch, A. Nayeem, and J.H. Freed.
J. Phys. Chem. 88, 3454-3465 (1984).
<doi: 10.1021/j150660a018>
Publication #126
|
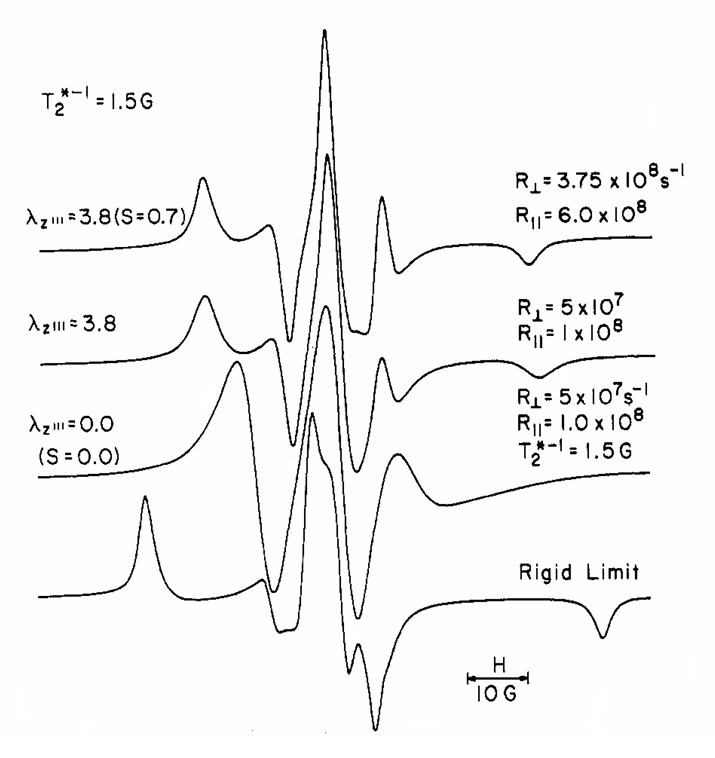
|
|
ABSTRACT: ESR spectra from protein-containing lipid dispersions have been interpreted in the past primarily in terms of two nitroxide species related respectively to "fluid" and "immobile" phospholipid environments. In this report we consider interpretations based primarily on a single type of lipid, for the two cases of spin-probe-doped membranes and of chemically labeled membrane proteins. The doped membranes are viewed as bilayer fragments with considerable local order but with macroscopic disorder of such fragments in the dispersion. In this "one-site" model all lipids are similar. They are fluid and oriented in their local environment. This model does not require a second species immobilized by contact with the proteins. The labeled proteins are considered as very slowly reorienting macromolecular complexes such that the dynamic effects on the ESR spectrum arise mainly from faster internal processes. The importance of, and the potential inherent in, detailed spectral simulation based on well-conceived models are emphasized by illustrating the great range of spectral line shapes that these models can yield with suitable parameters. Simulations are compared with some recent experimental examples previously interpreted in terms of a two-site model. Reasonably good results are obtained for spin-probe-doped membranes when allowance is made for local ordering as well as for some distortion of the alkyl chain ends. Such effects are modeled by introducing an ordering potential λ and a diffusion tilt angle Ψ which describes the tilt of the nitroxide moiety relative to the rest of the alkyl chain. The effects of adding protein or of lowering the temperature are modeled by increasing the local ordering while decreasing somewhat the motional rates with some increase in alkyl chain distortions. In the case of spin-labeled membrane proteins, the model of very anisotropic rotation, in which the side chain containing the nitroxide is rotating more rapidly than the protein about an effective axis tilted relative to the N-O axis, is found to account for the extra splittings with just a single-site model.
|
|
|
Two-Dimensional Electron Spin Echo Spectroscopy and Slow Motions
G.L. Millhauser and J.H. Freed.
J. Chem. Phys. 81, 37-48 (1984).
<doi: 10.1063/1.447316>
Publication #125
|
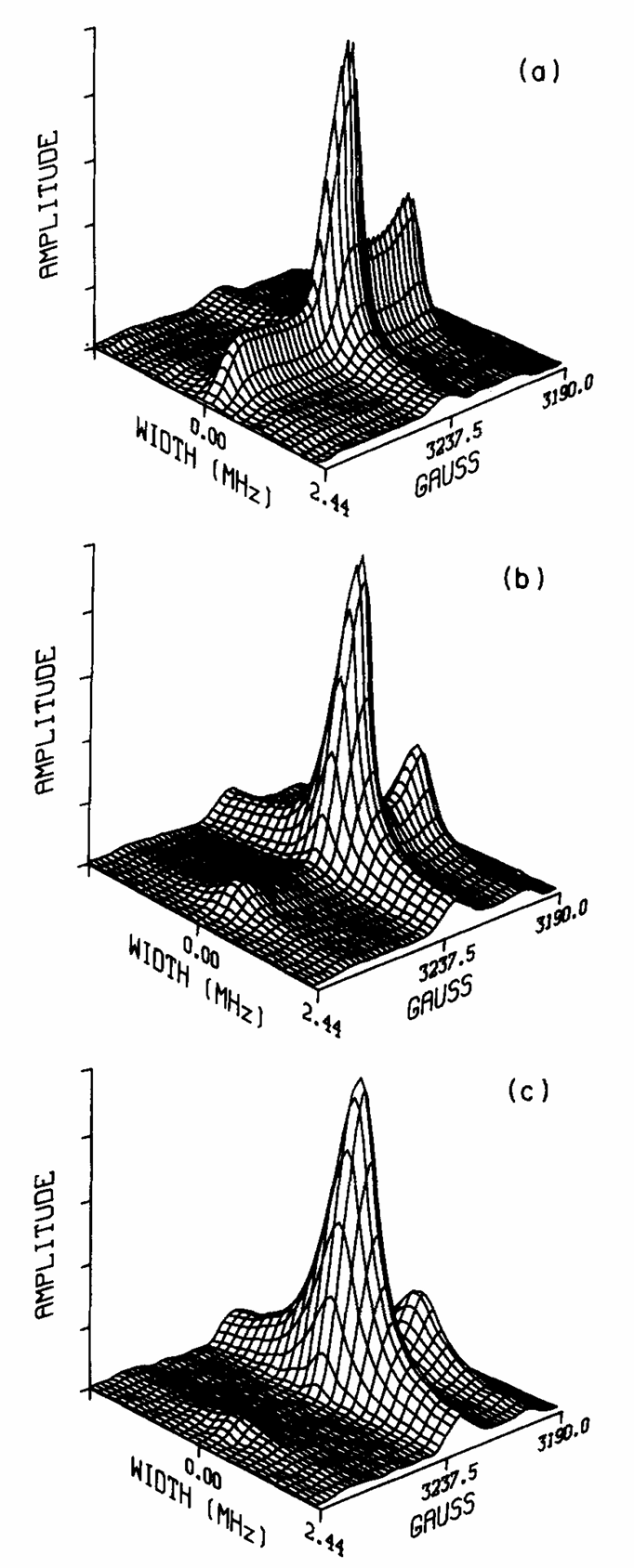
|
|
ABSTRACT: A technique is described for the study of slow molecular motions that employs two-dimensional electron spin echo (2D-ESE) spectroscopy. An "echo-induced ESR spectrum" is obtained with a two pulse 90° τ̱ 180° sequence by recording the echo height as the dc field is swept. A series of sweeps with different τ are Fourier transformed to yield a two-dimensional spectrum comprised of an echo-induced spectrum along the x axis and the homogeneous widths along the y axis. A convenient theoretical analysis appropriate for slow motions is given and it is shown that the 2D spectrum may be regarded as a graph of the widths vs the resonance positions of the individual "dynamics spin packets" that constitute the spectrum. An experimental study of the spin probe Tempone in 85% glycerol∕H2O solvent for slow motions (i.e., rotational correlational times of order 10−5–10−6 s) shows variations in homogeneous widths (i.e., T2−1) over the spectrum. This is analyzed in terms of canonical models of rotational reorientation—Brownian, free, and jump diffusion, and it is found that the variations of T2−1 across the spectrum show features similar to a Brownian model. A special normalized contour plot of the 2D spectra is introduced which clearly emphasizes the variation of T2−1 across the spectrum. The enhanced information content of these 2D spectra is discussed, and it is shown how 2D-ESE spectroscopy is both an improvement on and a complement to cw techniques for studying motional dynamics. Experimental extensions and limitations are also discussed.
|
|
|
Observation of Nuclear Spin Waves in Spin Polarized Atomic Hydrogen Gas
B.R. Johnson, J.S. Denker, N. Bigelow, L.P. Lévy, J.H. Freed, and D.M. Lee.
Phys. Rev. Lett. 52, 1508-1511 (1984).
<doi: 10.1103/PhysRevLett.52.1508>
Publication #124
|
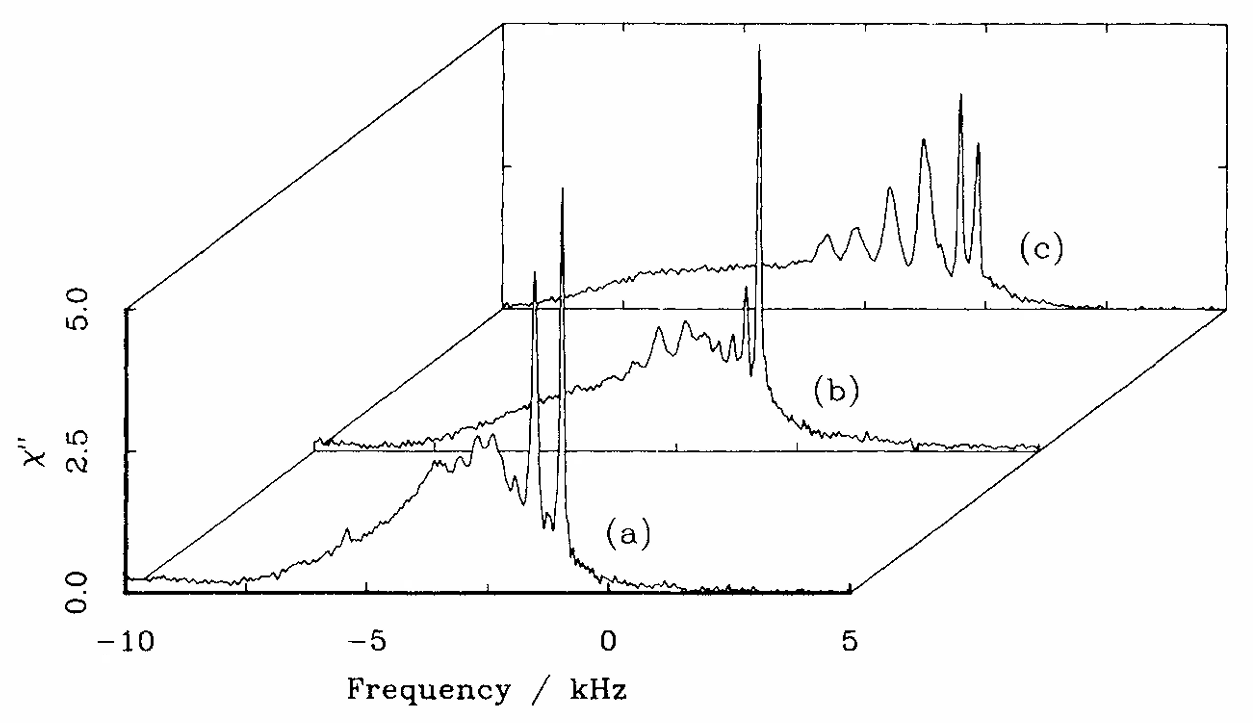
|
|
ABSTRACT: We have observed narrow, distinct resonances in the NMR spectrum of dilute spinpolarized atomic hydrogen gas (n˜1016 atoms∕cm3). The dependence of the observed spectra on temperature, density, polarization, and magnetic field gradient is consistent with theoretical predictions for spin-wave excitations damped by diffusion. We have measured the parameter μ, which is a measure of the importance of exchange effects in spin-transport processes, and the diffusion coefficient D0, both of which are in reasonable agreement with theory.
|
|
|
The Oscillatory Nature of Polarization Evolution in CIDEP
U. Eliav and J.H. Freed.
J. Phys. Chem. 88, 1277-1280 (1984).
<doi: 10.1021/j150651a005>
Publication #123
|
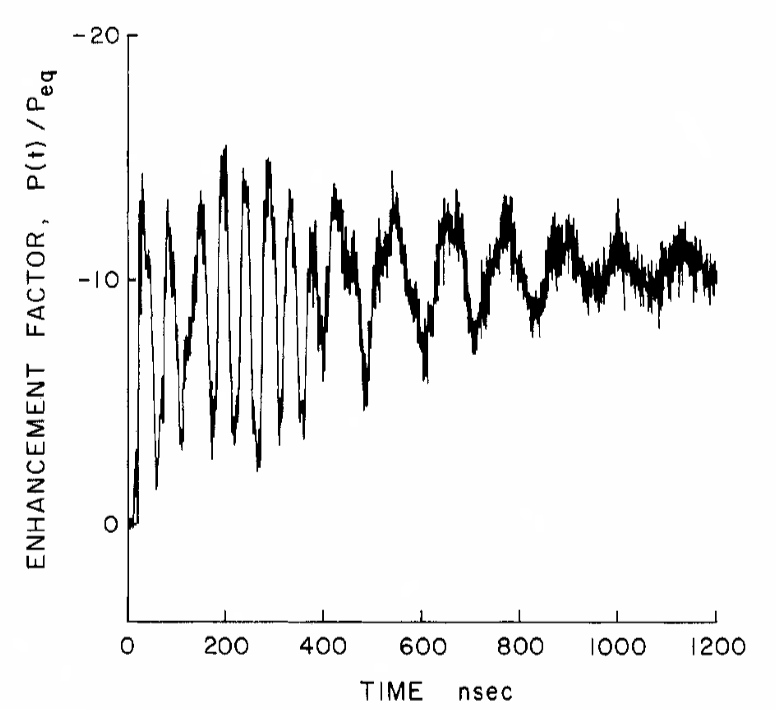
|
|
ABSTRACT: An unusual and striking pattern of oscillations in the time evolution of the polarization has been observed in a time-resolved CIDEP study of photoelectrons generated in high concentration Rb∕THF solutions. It is suggested that such oscillations may be characteristic of a (geminate) radical-pair mechanism when observed under sufficient time resolution.
|
|
|
ELDOR Spin Echoes and Slow Motions
J.P. Hornak and J.H. Freed.
Chem. Phys. Lett. 101, 115-119 (1983).
<doi: 10.1016/0009-2614(83)87353-9>
Publication #122
|
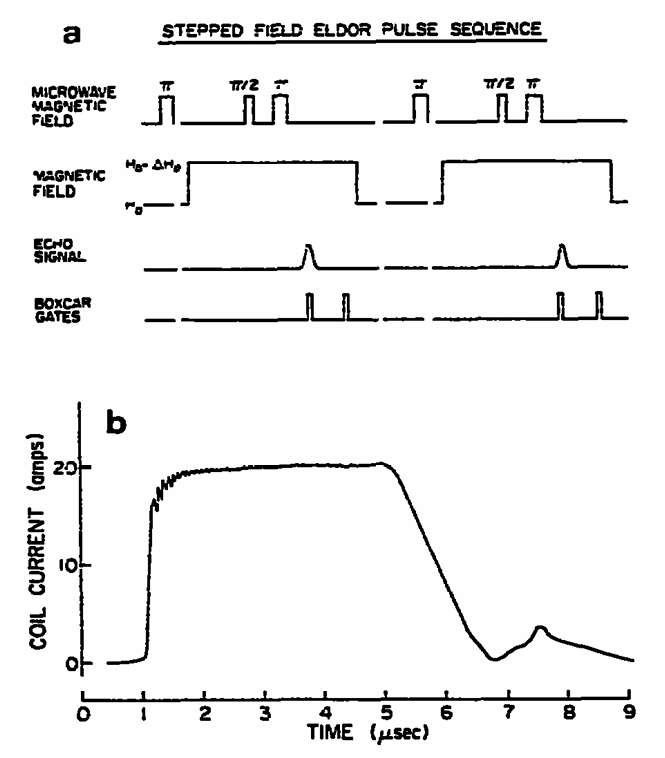
|
|
ABSTRACT: It is shown how an ELDOR technique based upon spin echoes and rapid stepping of the magnetic field may be employed to measure rotational correlation times, τR for very slow motions. Experiments on PD-Tempone in 85% glycerol∕ D2O at low temperatures led to τR values of 10−4 to 10−5 s obtained with a simple analysis of the data.
|
|
|
Electron Spin-Echo Studies of Relaxation Times in Lithium-Methylamine Solutions
C.J. Page, G.L. Millhauser, P.P. Edwards, J.H. Freed, and M.J. Sienko.
J. Phys. Chem. 88, 3785-3789 (1984).
<doi: 10.1021/j150661a021>
Publication #121
|
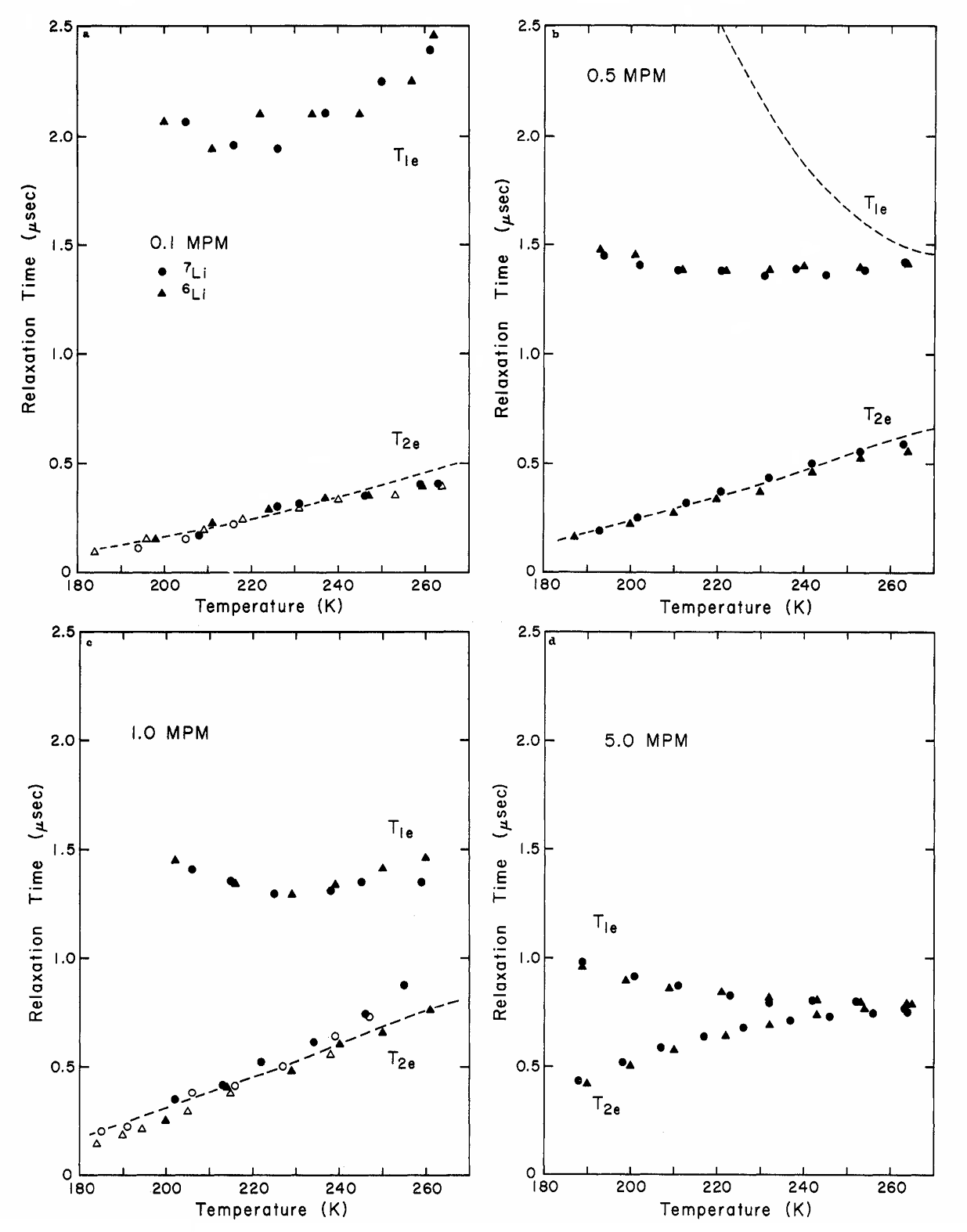
|
|
ABSTRACT: The electron spin relaxation times T1 and T2 of lithium-6- and lithium-7-methylamine solutions have been measured by electron spin-echo techniques over the temperature range 200-260 K. T1 and T2 are not equal in the concentration range studied (0.1-5.0 MPM); their values differ most at low concentrations and low temperatures. No significant lithium isotope effect is observed. The accepted relaxation mechanism for dilute metal-ammonia solutions (motional modulation of the electron-14N hyperfine interaction) cannot alone account for the observed relaxation times in lithium-methylamine solutions.
|
|
|
Transverse Viscous Forces in Carr Walls and Possible Dynamic Consequences
D. Igner and J.H. Freed.
Mol. Cryst. Liq. Cryst. 101, 301-313 (1983).
<doi: 10.1080/01406568308072538>
Publication #120
|
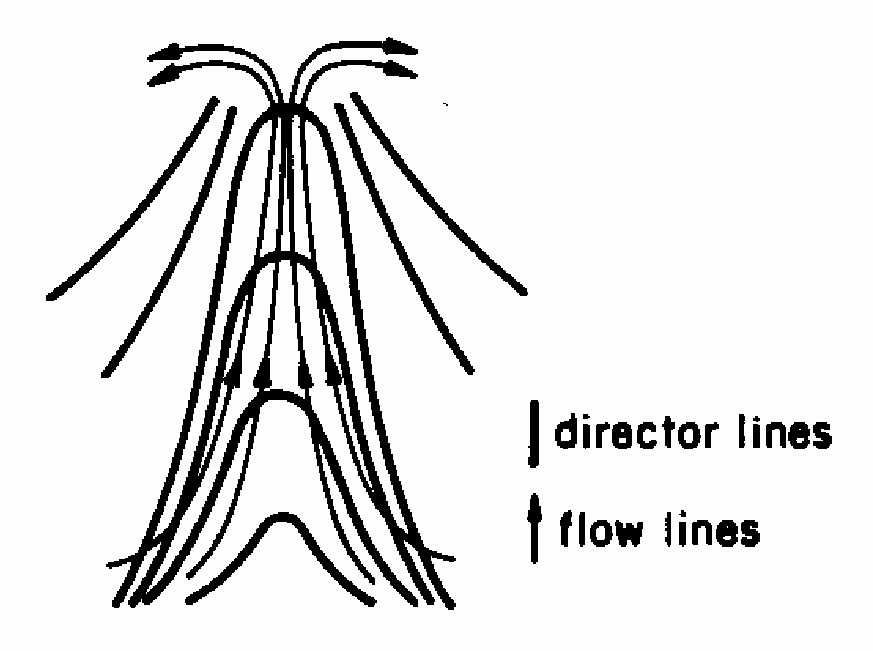
|
|
ABSTRACT: Expressions for the anisotropic viscous forces transverse to a nematic flow confined in a straight splay-bend wall are derived under simplifying assumptions. Some possible dynamic consequences, such as longitudinal splitting of the walls and movement of asymmetric walls in electro-hydrodynamic instabilities, are suggested. The relevance to recent experimental observations is discussed.
|
|
|
Stochastic Modeling of Molecular Dynamics
J.H. Freed.
In Stochastic Processes-Formalism and Applications, G.S. Agarwal and S. Dattagupta, Eds.
Lecture Notes in Physics 184, 220-225 (1983).
Publication #119
|
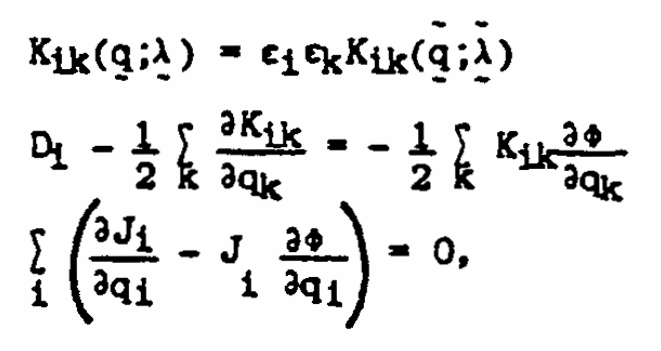
|
|
INTRODUCTION: We outline here a method, due to Stillman and Freed, for the stochastic modeling of the non-Markovian many-body features of diffusing molecules. We believe this approach is a particularly useful one in that it introduces, in a transparent manner, the basic physics of the relevant degrees of freedom and their couplings, and it is not restricted to linear transport laws. Furthermore, the stochastic features of the bath variables are introduced in a simple and physically transparent fashion. Also, this approach permits consideration of either equilibrium or non-equilibrium dynamics in that the modeling allows for physically relevant choice of equilibrium or stationary-state, and it subjects the expressions to the constraints of detailed balance with respect to this state.
In this method the set ot relevant dynamical variables is first augmented with stochastic bath variables which are assumed to obey simple Markovin laws. The augmented set then represents a multidimensional Markov process which obeys a stochastic Liouville equation (SLE) that is, in general, incomplete because it ignores the back reaction of the molecule (i.e. the relevant dynamical variables) on the bath variables. The back reaction effects are incorporated into the model by adding terms to the SLE which are obtained by the constraints required for detailed balance. The resulting augmented Fokker-Planck equation (AFPE) properly describes relaxation to thermal equilibrium, and, for the appropriate limiting conditions, reduces to a classical Fokker-Planck equation. Augmented Langevin equations (ALE) may readily be obtained from the AFPE, and because of the constraint of detailed balance, they automatically obey the fluctuation-dissipation theorem.
|
|
|
Multipulse Sequences in Electron-Spin Echoes
U. Eliav and J.H. Freed.
Rev. Sci. Instr. 54, 1416-1417 (1983).
<doi: 10.1063/1.1137235>
Publication #118
|
|
|
ABSTRACT: A method is described whereby Carr–Purcell (CP) multipulse sequences may be generated at a rate rapid enough for electron-spin echoes ESE and in a manner consistent with constraints of typical ESE spectrometers. Both Carr–Purcell–Meiboom–Gill and alternating phase-CP sequences may be produced. It is shown how such sequences may be utilized (1) to increase the rate of data collection, which is especially important in laser photo-induced experiments, and (2) to study slow motions. They are respectively illustrated with results on transient spin-polarized photoelectrons and electrons trapped in irradiated quartz.
|
|
|
Anisotropic Molecular Motion of Spin-Labeled Poly(Methyl Methacrylate) Detected by ESR
M. Shiotani, J. Sohma, and J.H. Freed.
Macromolecules 16, 1495-1499 (1983).
<doi: 10.1021/ma00243a016>
Publication #117
|
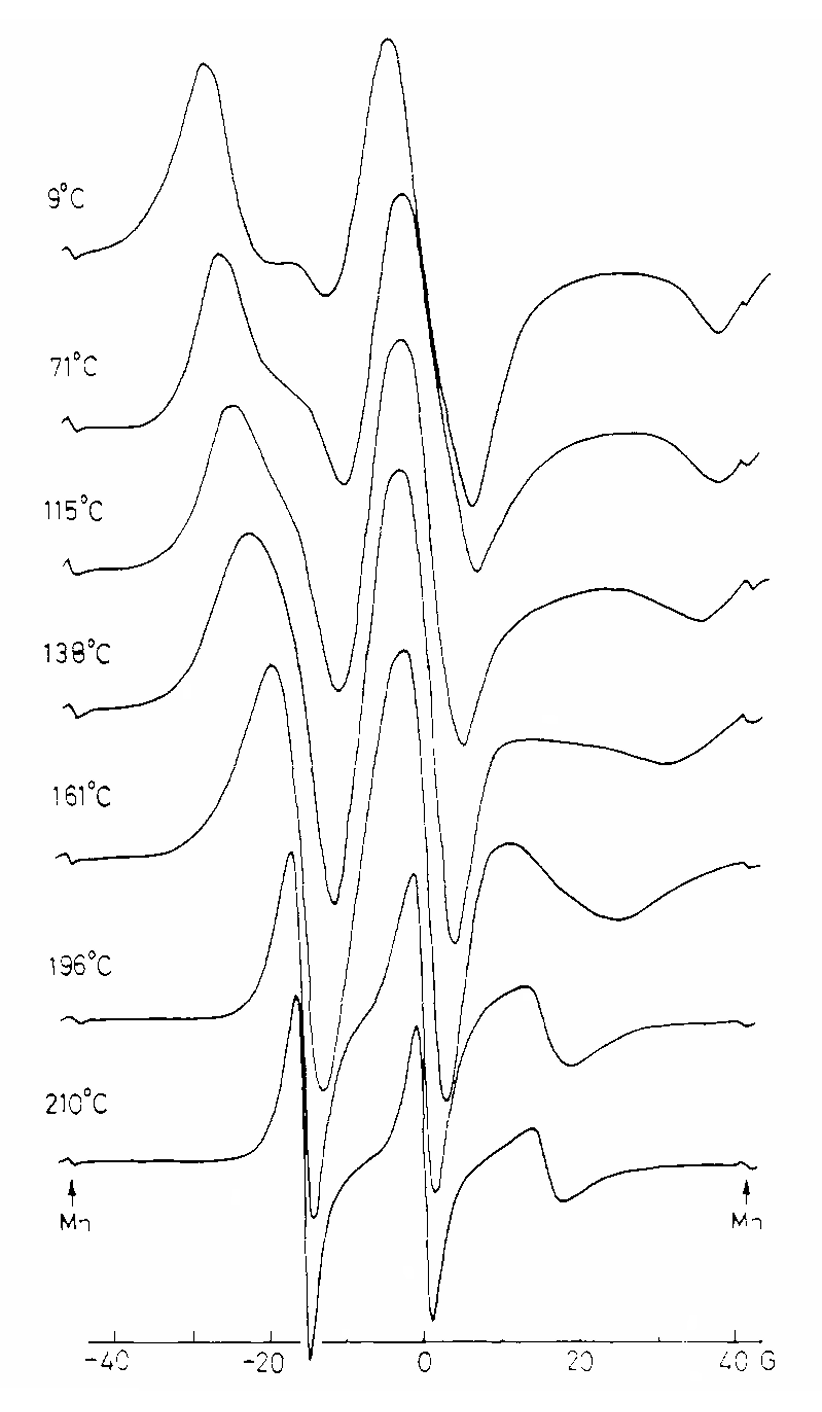
|
|
ABSTRACT: Anisotropic rotational motion of the side chain in poly(methyl methacrylate) (PMMA) is inferred from the detailed analysis of the ESR spectrum of spin-labeled PMMA. Rotation about the axis parallel to the N-O bond of the nitroxide label bonded at the end of the ester side chains was twice as fast as that about the axes perpendicular to this bond in the incipient motional narrowing region. This axially symmetric rotation is discussed in terms of the molecular motions of PMMA.
|
|
|
Quadrupole Echo Study of Internal Motions in Polycrystalline Media
L.J. Schwartz, E. Meirovitch, J.A. Ripmeester, and J.H. Freed.
J. Phys. Chem. 87, 4453-4461 (1983).
<doi: 10.1021/j100245a026>
Publication #116
|
|
|
ABSTRACT: Quadrupole echo spectra of polycrystalline trimethylsulphoxonium-d9 iodide (TMSI-d9) and hexamethyl-benzene-d18 (HMB-d18) in the slow- and fast-motional temperature regions are analyzed via a dynamic line-shape formalism which incorporates the stochastic Liouville operator. Both molecules are found to exhibit two approximately independent motional modes: methyl-group rotation, and discrete jumps about a single axis. For TMSI-d9, this latter mode consists of 3-fold jumps about an axis tilted 67.5 ± 1.0° with respect to the S-C axis, with an activation energy EA = 11.1 ± 0.9 kcal/mol and an inverse frequency factor τ0 ≃ 6 × 10−13 s, while HMB-d18 reorients about its hexad axis with EA = 5.3 ± 0.2 kcal/mol and τ0 ≃ 2 × 10−12 s. Simulations based on the Fourier transforms of calculated free induction decays (instead of calculated echoes) are inadequate for much of the temperature ranges studied.
|
|
|
On Cooperative Modes of Reorientation in Liquid Crystals
G.P. Zientara and J.H. Freed.
J. Chem. Phys. 79, 3077-3089 (1983).
<doi: 10.1063/1.446138>
Publication #115
|
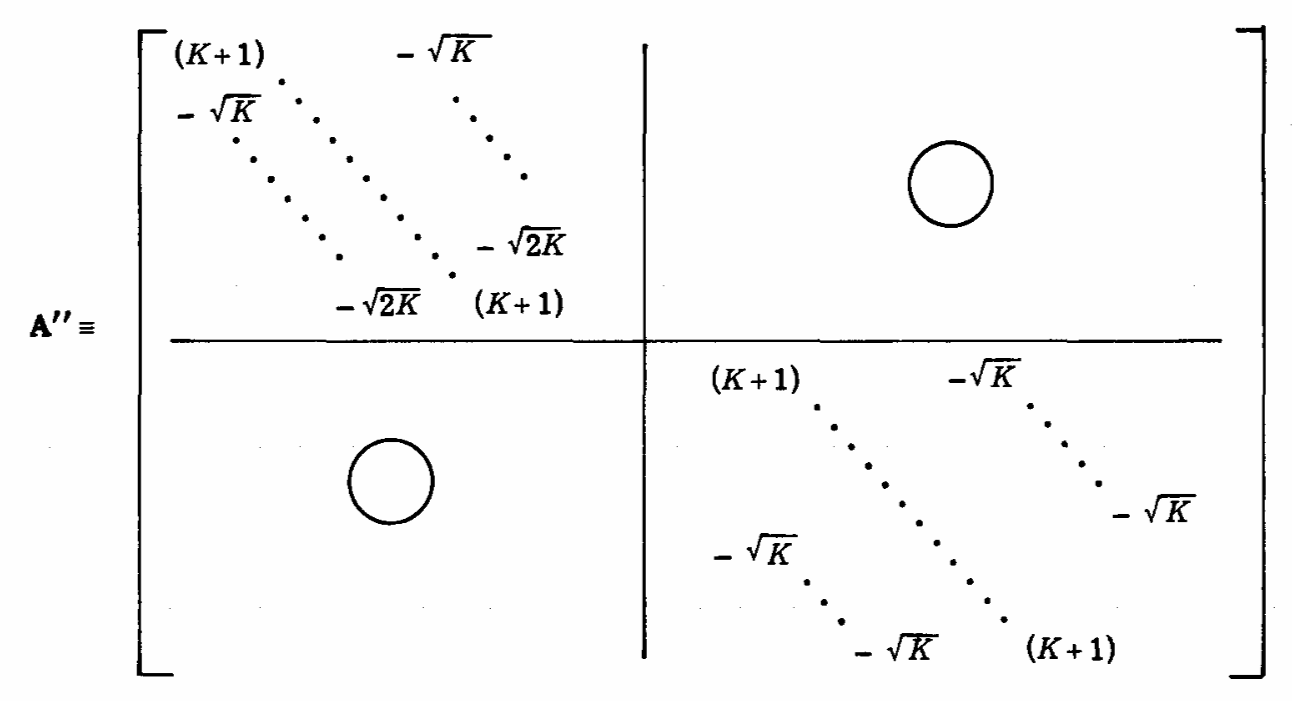
|
|
ABSTRACT: The role of cooperative modes of reorientation in liquid crystals for the spectral densities of importance in magnetic resonance relaxation is examined from the viewpoint of microscopic models that can be solved analytically. These include linear arrays of rods in an applied external field whose orientations are strongly coupled and whose motion is described by overdamped and coupled diffusion equations. Also considered are Cayley-tree models, which, at least for nearest neighbors, can mimic higher dimensions than one. All these models lead to the result that the qth normal mode of relaxation has a decay rate that depends upon q as sin2(qa∕2), where a is the lattice spacing. Using simple scaling, it is argued that if a is replaced by a′≡La, so our model consists of blocks of L rodlike molecules with renormalized parameters, then molecular detail becomes less important and the model arrays become more physically representative. These results are then utilized to extend the standard hydrodynamic theory for long wave vectors q to short wave vectors approaching molecular dimension. The spectral densities obey scaling (i.e., are nearly independent of a′) and are nearly identical to the hydrodynamic result for lower frequencies. However, the new results do show small departures at the higher frequencies usually studied by NMR.
|
|
|
Molecular Orbital Study of O2- Adsorbed on Titanium Ions on Oxide Supports
K. Tatsumi, M. Shiotani, and J.H. Freed.
J. Phys. Chem. 87, 3425-3429 (1983).
<doi: 10.1021/j100241a015>
Publication #114
|
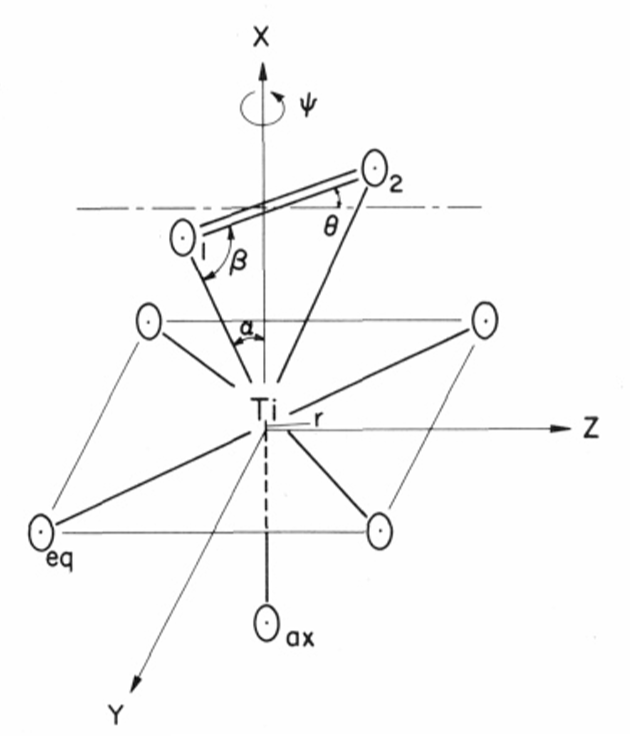
|
|
ABSTRACT: An extended Hückel analysis of O2− adsorbed on Ti ions on oxide supports is presented. The potential energy surface is obtained for deformations between the two limiting structures of the O2− parallel to the surface and end-on. A stable conformation is found for the parallel form, consistent with recent ESR results, and there is a maximum in energy between the two limiting structures. The observed near equality of the ESR hyperfine splittings of the two O atoms is found to be consistent with a small tilting of the O2 axis from the parallel conformation (i.e., <10°). A low barrier for planar rotation of O2− about the axis perpendicular to the surface is also found, consistent with the small activation energy obtained from ESR line-shape analysis. The predicted g-tensor components allow unambiguous assignment of the ESR results, and a discussion is given on how variations in the extended Hückel parameters improve agreement with experiment.
|
|
|
NMR-Induced Recombination of Spin Polarized Hydrogen
B. Yurke, J.S. Denker, B.R. Johnson, N. Bigelow, L.P. Lévy, D.M. Lee, and J.H. Freed.
Phys. Rev. Lett. 50, 1137-1140 (1983).
<doi: 10.1103/PhysRevLett.50.1137>
Publication #113
|
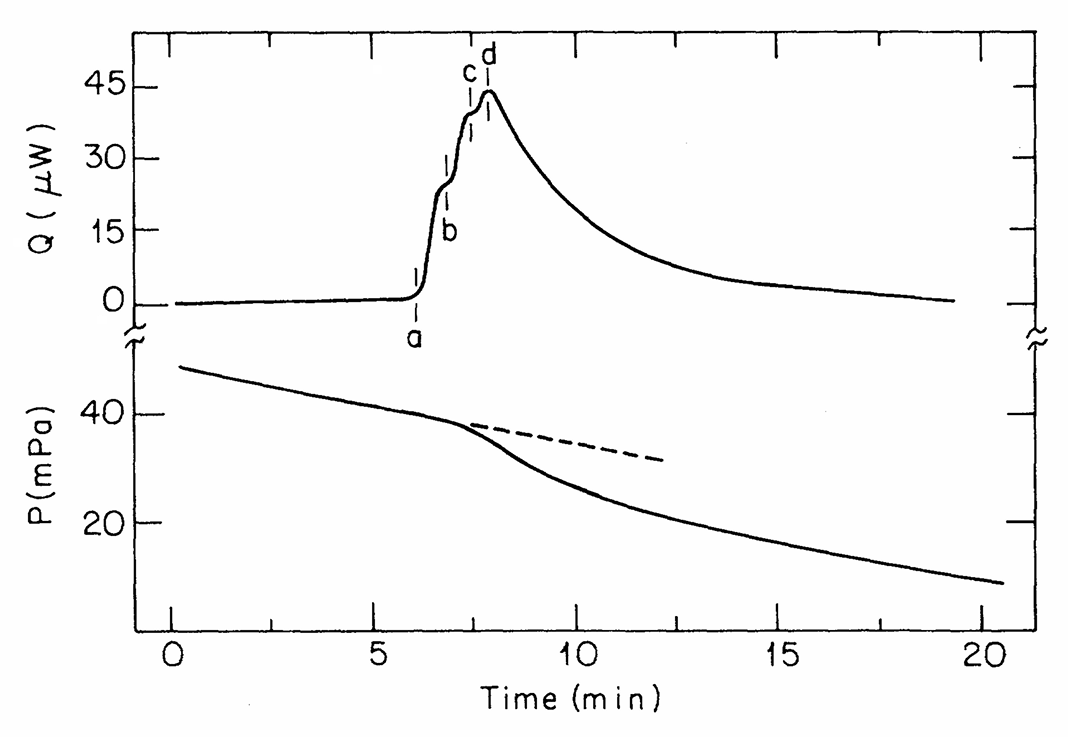
|
|
ABSTRACT: The presence of an inverted population of H↓ with very high electronic and nuclear spin polarization is confirmed with a new barometric-NMR method and by free-induction decay. With this new technique, the temperature dependence for the ratio of the two state-dependent recombination rate constants is measured. The rate of spin relaxation in the gas agrees with the latest theory, but relaxation on the surface is much faster than predicted, and there is an important surface one-body relaxation process.
|
|
|
Analysis of Electron Spin Echoes by Spectral Representation of the Stochastic Liouville Equation
L.J. Schwartz, A.E. Stillman, and J.H. Freed.
J. Chem. Phys. 77, 5410-5425 (1982).
<doi: 10.1063/1.443791>
Publication #112
|
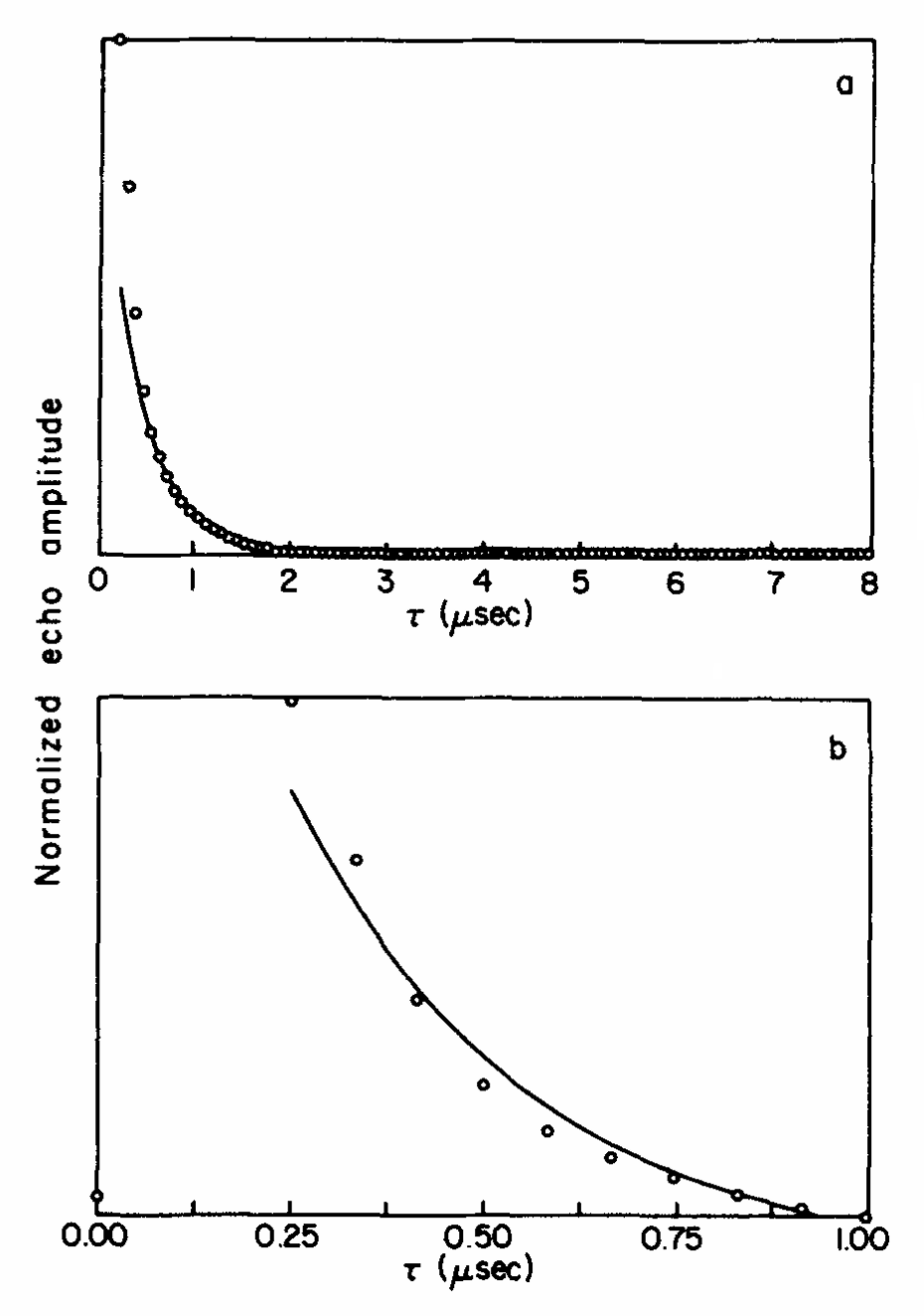
|
|
ABSTRACT: Motional effects can easily be incorporated into calculated spin echo decay envelopes by idealizing the effects of the pulses and by using the stochastic Liouville equation (SLE) to govern the time dependence of the density matrix between pulses. The spectral representation of the 90°-τ-180°-τ envelope is: Σl,mal,mexp[−(Λl+Λ*m)τ], where Λl is the lth eigenvalue of the SLE matrix, and al,m are products of relevant components of eigenvectors. The long time (large τ) phase memory time Tm∞ is equal to (Re Λ1)−1, where Λ1 is the smallest eigenvalue. For an axially symmetric g-tensor case in the slow motional region, Tm∞∕τR ≃ 3∣FτR∣−1∕2, 2∣FτR∣−1∕3, and 1, for Brownian, free, and jump diffusion models, respectively, with F≡2βeH0(g∥−g⊥)∕3ℏ and τR the rotational correlation time. Analogous results hold for nitroxides having nuclear hyperfine tensors. The echo results are compared with simple methods of measuring τR from cw spectra. The overall shape of the echo envelopes in the slow motional region is exp(−bF2τ3∕τR) for very short times and exp(−2τ∕Tm∞) for the longer times. Carr–Purcell (CP) sequences suppress the initial exponential in τ3 and increase the phase memory times as a function of decreasing τ. Detailed analysis of CP sequences can provide information on motional model. The analysis of motional averaging of nuclear modulation effects by our method is also given and it is pointed out that this approach may be useful in studying very slow motions. Despite the simplicity of the methods, we are able to approximately fit experimental data from the Tempone∕glycerol–water system in both fast and slow motional regions.
|
|
|
Electron-Spin Relaxation and Molecular Dynamics in Liquids II. Density Dependence
S.A. Zager and J.H. Freed.
J. Chem. Phys. 77, 3360-3375 (1982).
<doi: 10.1063/1.444278>
Publication #111
|
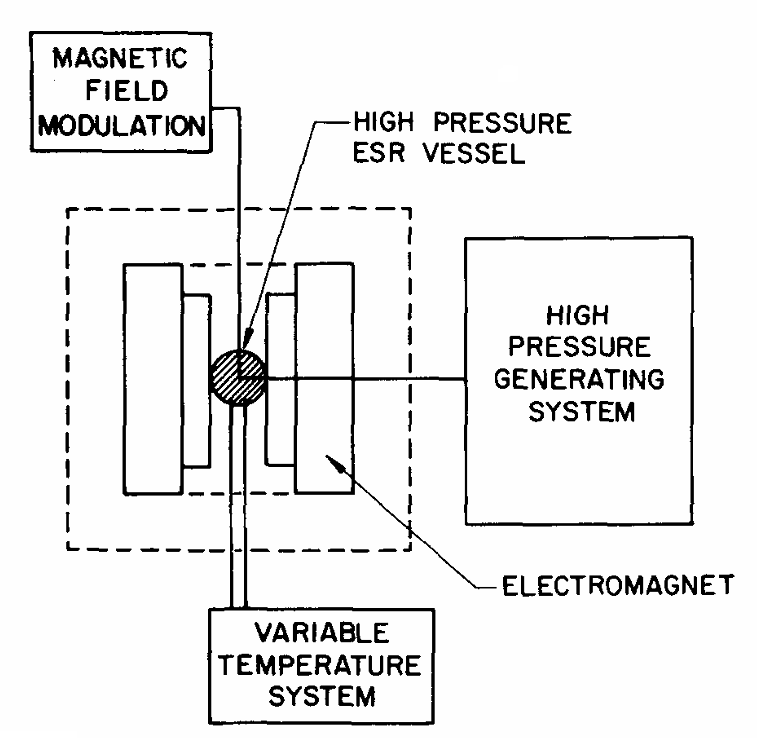
|
|
ABSTRACT: A pressure-dependent ESR relaxation study of the probe PD-tempone dissolved in toluene-d8 is described. Extensive results on rotational relaxation, in which τR(T,P) is varied over more than two orders of magnitude, are presented. These results are found to be inconsistent with a simple Stokes–Einstein type η∕T behavior modified with a nonzero intercept. However, the data were successfully fit by the empirical form τR=cηβ(ρ−ρ̃)∕T
where β is the isothermal compressibility, c is a constant, and ρ̃ is an (empirical) reference density whose inverse can be thought of as an "expanded volume". Thus, ρ−ρ̃ is a measure of the strength of the anisotropic intermolecular interactions acting on the probe, while β−1 may be thought of as a measure of the total intermolecular interactions. These results for a solute of molecular size somewhat greater than that of the solvent molecules exhibit some differences when compared to a previous NMR study on neat toluene-d8. The ε parameter introduced by Freed and co-workers to fit their nonsecular spectral densities is found to be independent of temperature and pressure, and it is pointed out that this could be consistent with an intermolecular fluctuating torque model.
|
|
|
Electron-Spin Relaxation and Molecular Dynamics in Liquids I. Solvent Dependence
S.A. Zager and J.H. Freed.
J. Chem. Phys. 77, 3344-3359 (1982).
<doi: 10.1063/1.444304>
Publication #110
|
|
|
ABSTRACT: Temperature-dependent ESR relaxation studies of PD-Tempone in a variety of solvents ranging from weakly interacting hydrocarbons to strongly interacting D2O are described. The empirical molar transition energy scale ET, which is a good measure of specific solvent polarity, is found to lead to a useful interpolation method for estimating the solvent dependence of the magnetic tensors required for accurate spin-relaxation studies. The temperature-dependent results are generally well-represented by a modified Stokes–Einstein type of η∕T dependence for τR, the rotational correlation time. The ε parameter introduced by Freed and co-workers as an empirical correction to the nonsecular Debye spectral densities is found to correlate with ET, such that it is largest for the weakly interacting hydrocarbon solvents and approaches the Debye limit of ε = 1 for D2O solvent. This is discussed in terms of approximate fluctuating torque and slowly-relaxing local structure models, and the former appears more consistent with experiment. It is also pointed out that ε≳1 would result from the phenomenological feature of viscoelasticity.
|
|
|
Electron-Spin Relaxation and Ordering in Smectic and Supercooled Nematic Liquid Crystals
E. Meirovitch, D. Igner, E. Igner, G. Moro, and J.H. Freed.
J. Chem. Phys. 77, 3915-3938 (1982).
<doi: 10.1063/1.444346>
Publication #109
|
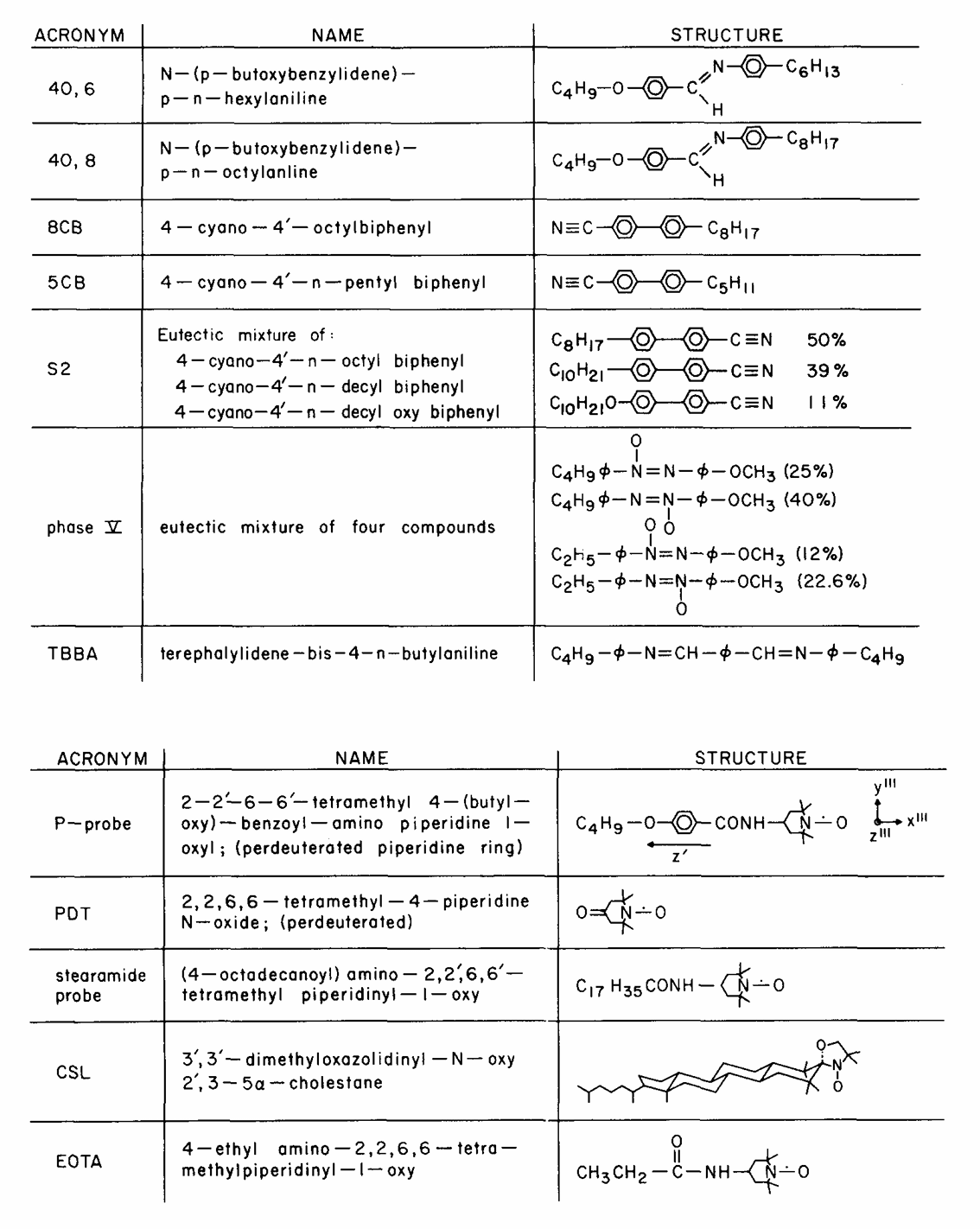
|
|
ABSTRACT: We report on careful line shape studies of slow motional and orientation dependent ESR spectra of a deuterated liquid-crystal-like spin probe dissolved in a benzilidene-derivative (40,6) and in cyanobiphenyl derivative (S2 and 5CB) liquid crystals. The simulation of the ESR spectra is based on the Lanczos algorithm recently applied by Moro and Freed in a general and efficient formulation of slow motional and ordering effects on ESR line shapes. With 40,6 which exhibits monolayer smectic phases, we find that the main change in the spin relaxation upon passing from the nematic to the smectic A phase consists of changes occuring in ordering attributable to packing forces on functional groups. Such ordering effects appear to be further enhanced in the SB phase with consequent alterations in dynamics. With S2, which exhibits an interpenetrating bilayer smectic A phase, we find unusual ESR spectra in that phase which may be simulated on the basis of a model of cooperative distortions static on the ESR time scale, and superimposed on individual molecular reorientation. This mode is interpreted as a collective chain distortion which affects the orientational distribution of the piperidine ring of the spin probe. A similar phenomenon is observed in the supercooled nematic phase of 5CB, which is aligned by an electric field, and evidence is also found that the reorientational dynamics of this ring are affected by interaction with local cooperative modes in the liquid crystal (i.e., a SRLS mechanism previously proposed by Freed and co-workers). Some microscopic characteristics of liquid crystals revealed by this and previous ESR spin probe studies are discussed.
|
|
|
Electrohydrodynamic Instabilities Observed in a Nematic Phase under Oblique Boundary Conditions
D. Igner and J.H. Freed.
J. Chem. Phys. 76, 6095-6119 (1982).
<doi: 10.1063/1.443014>
Publication #108
|
|
|
ABSTRACT: Electrohydrodynamic instabilities in the nematic phase of Merck "Phase V" with oblique boundary conditions were optically observed with a polarizing microscope in 25–100 μm "sandwich" cells. Oblique anchoring of the nematic was achieved by oblique evaporation of SiO on the plates. Two types of cells were used having the respective in-plane projection of the direction of evaporation on the two plates either parallel (p-type cells), or antiparallel (a-type cells). The low voltage dc instability observed for the p-type cells forms in an almost regular hexagonal pattern. By gradually increasing the voltage, the dc instability observed for the a-type cells forms at first as flows which originate at order disturbances created at imperfections in the SiO coating. Voltage increase causes these flows to detach themselves from the places of the imperfections and move solitarily. The moving flows are associated with what appears to be moving tilt inversion deformations (of splay-bend type) extending from the central part of the flow to some distance from it. When the voltage is further increased, a repeated process of replication of the flows, occurring on the associated tilt inversion deformations, leads to the creation of a periodic grid of moving flows. Other observed types of static and dynamic patterns under ac and dc excitation are reported, in particular: different types of cross rolls (ac conduction regime); variations of a pattern of what appears to be walls associated with flows, exhibiting an approximate wave number dependence on the electric field k˜E; a striped pattern associated with what appears to be twist walls and the propagating interference patterns associated with their oscillations; a toroidal flow (sometimes associated with closed inversion walls) which creates and caries along closed nematic threads (dc regime); a polygonal grid of turbulent flows (dc regime); a flow pattern correlated with the movement of the moving chevron pattern; a cellular fast turn-off pattern related to the chevron pattern. This cellular pattern appears at first as moving snakelike regions in the chevron pattern which are bordered by disclination lines. Some features of dark, spotlike figures appearing on the chevron pattern are described. Preliminary interpretations of some of the observations are offered.
|
|
|
Photoelectric Determination of Work Function Via CREMSEE Enhancement. Photo-CREMSEE
M. Nilges and J.H. Freed.
Chem. Phys. Lett. 85, 499-504 (1982).
<doi: 10.1016/0009-2614(82)80345-X>
Publication #107
|
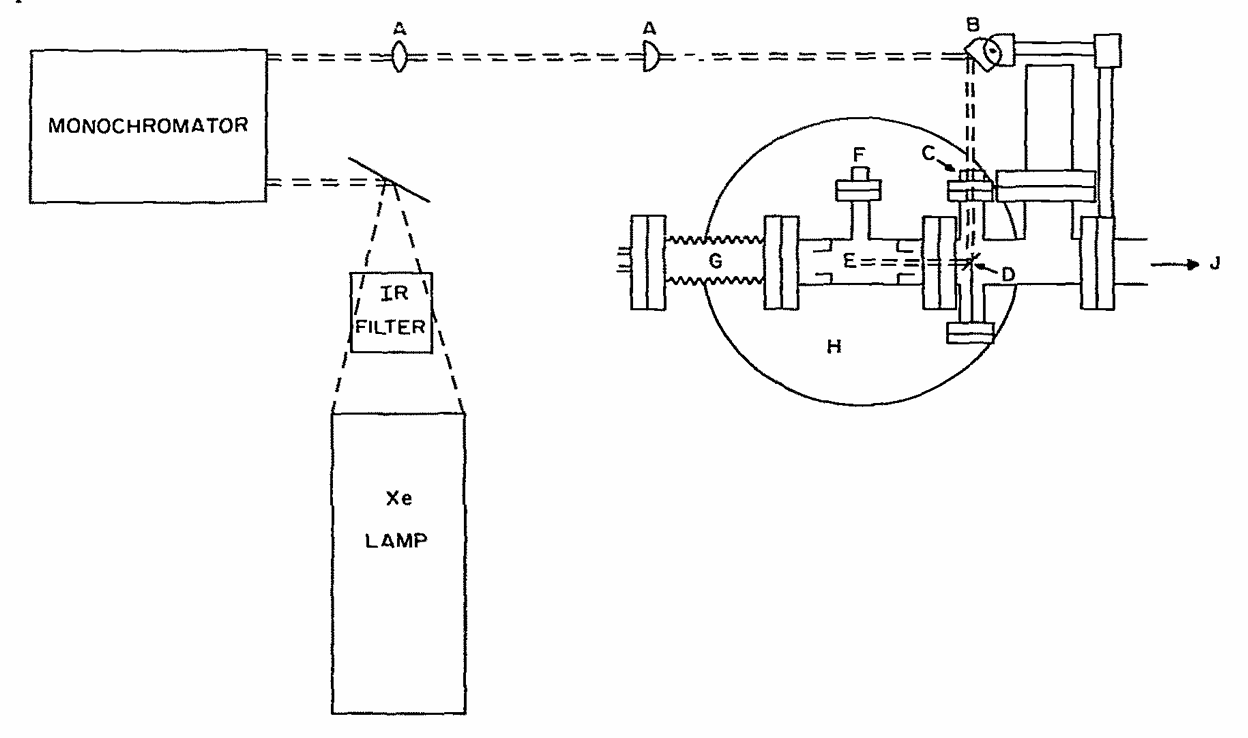
|
|
ABSTRACT: CREMSEE (cyclotron resonance from microwave-induced secondary electron emission), operated at a microwave power level just below threshold for self-sustaining signals may be used as an election-amplifier and detector of photoetectrons. A determination of the work function of air-oxidized Ag on the interior surface of a UHV-microwave cavity yields 4.06 ± 0.05 eV.
|
|
|
A Quantum Stochastic Theory for Nonadiabatic Processes in Condensed Phases and on Surfaces
W.A. Wassam, Jr. and J.H. Freed.
J. Chem. Phys. 76, 6150-6169 (1982).
<doi: 10.1063/1.443018>
Publication #106
|
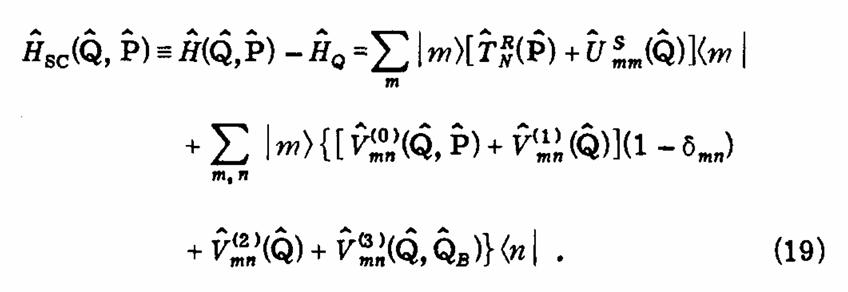
|
|
ABSTRACT: A quantum stochastic theory for nonadiabatic processes in condensed phases and on surfaces is given. The present theory is primarily concerned with processes involving the fragmentation of two-particle systems and/or the "collision" of two particles with internal structure, i.e., electronic, spin, and nuclear degrees of freedom. A fully quantum mechanical joint energy space–quantum phase space master equation for two-particle systems is presented. A simpler form of this equation emerges in the limit where the length of thermal spatial fluctuations along the relative coordinates is much greater than half the wavelength for thermal momentum fluctuations associated with relative motion. (This limit is equivalent to fulfilling a "thermal Heisenberg uncertainty" relation: Δp̄Δq̄≫ℏ∕2.) The simpler master equation is converted into a nonlinear quantum Fokker–Planck equation for nonadiabatic systems. Introducing simplifying assumptions, we obtain a quantun analog of the stochastic Liouville equation augmented with irreversible kinetic terms, which describes nonadiabatic transitions and the interruption of coherence in energy space at different points in quantum phase space. The final results are appropriately modified to incorporate the influence of applied time-dependent electric and magnetic fields. The present work provides a formal theoretical framework for examining the role of quantum effects that are neglected in "semiclassical" kinetic and "diffusional" theories for nonadiabatic processes.
|
|
|
A Quantum Stochastic Fokker-Planck Theory for Adiabatic Processes in Condensed Phases
W.A. Wassam, Jr. and J.H. Freed.
J. Chem. Phys. 76, 6133-6149 (1982).
<doi: 10.1063/1.443017>
Publication #105
|
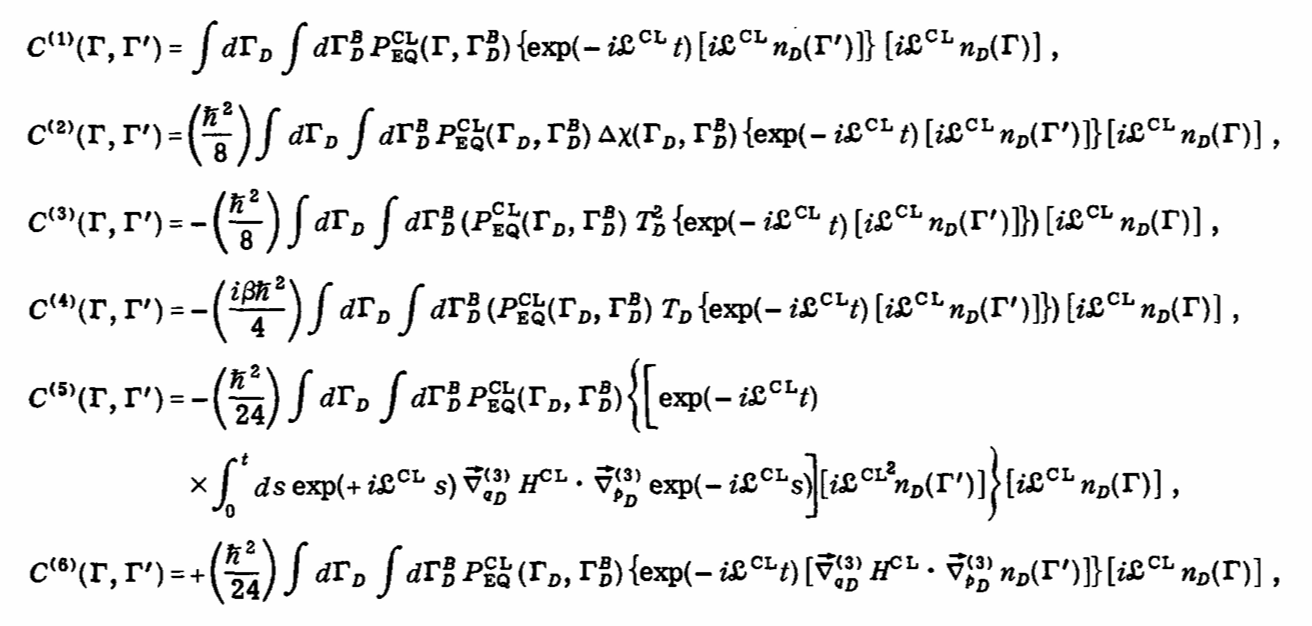
|
|
ABSTRACT: A maximum entropy approach to time-dependent phenomena is employed to construct a quantum stochastic theory for adiabatic processes in condensed phases. This theory is illustrated by considering the relative motion of a two-particle system interacting with a heat bath. We present a fully quantum mechanical phase space master equation, which is new. In the limit where the length of thermal spatial fluctuations along the relative coordinates is much greater than half the wavelength for thermal momentum fluctuations associated with relative motion, our master equation reduces to a simpler form, and the principle of detailed balance emerges. This limit is shown to be equivalent to fulfilling a "thermal Heisenberg uncertainty" relation: Δp̄Δq̄≫ℏ∕2. The simpler master equation is utilized in the development of a quantum mechanical nonlinear Fokker–Planck equation. For cases in which correlations involving spatial and momentum fluxes are short ranged, the nonlinear Fokker–Planck equation reduces to the usual linear form, with "collision" and "streaming" terms that contain quantum corrections to all orders in ℏ. The relationship between the classical limit of the present theory and Brownian motion theory based on the Chapman–Kolmogorov equation is established.
|
|
|
Nitroxides and Chemical Physics
J.H. Freed.
Finnish Chemistry 9, 50-51 (1982).
Publication #104
|
|
|
INTRODUCTION: The availability of stable nitroxide free radicals has proved a boon for the application of ESR to a number of fields in chemical physics.
We have been using small nitroxide spin probes for studying molecular dynamics in liquids, and have shown the kind of detailed information which one may obtain from ESR studies. In general, we have been exploring experimental evidence for the breakdown of simple Brownian motional models for rotational reorientation. One particular problem we have considered is the limitation
of the simple Stokes Law: τRSE = 4πηa3∕3kT for the rotational correlation time τR. Our previous results have all been consistent with a linear relation between τR and η∕T, although a variety of non-ESR experiments have implied deviations. We have completed spin relaxation studies on the system perdeuterated tempone (PDT) in toluene as a function of both temperature and pressure using previously reported methods, and we have obtained systematic deviation between τR and τRSE which could be represented in the form τR∕τRSE = A − BT + CT2 + DT∕o̢, where o̢ is the density, and A, B, C, and D are constants independent of T and o̢. These deviations represent the differences between the molecular details of reorientation and the hydrodynamic behavior of a Stokes-Einstein model. They appear to be inconsistent with recent molecular and hydrodynamic models. The very precise and detailed ESR results seem particularly useful for such studies.
|
|
|
Dynamics of Protein Domain Coalescence: 2
G.P. Zientara, J.A. Nagy, and J.H. Freed.
J. Phys. Chem. 86, 824-832 (1982).
<doi: 10.1021/j100394a046>
Publication #103
|
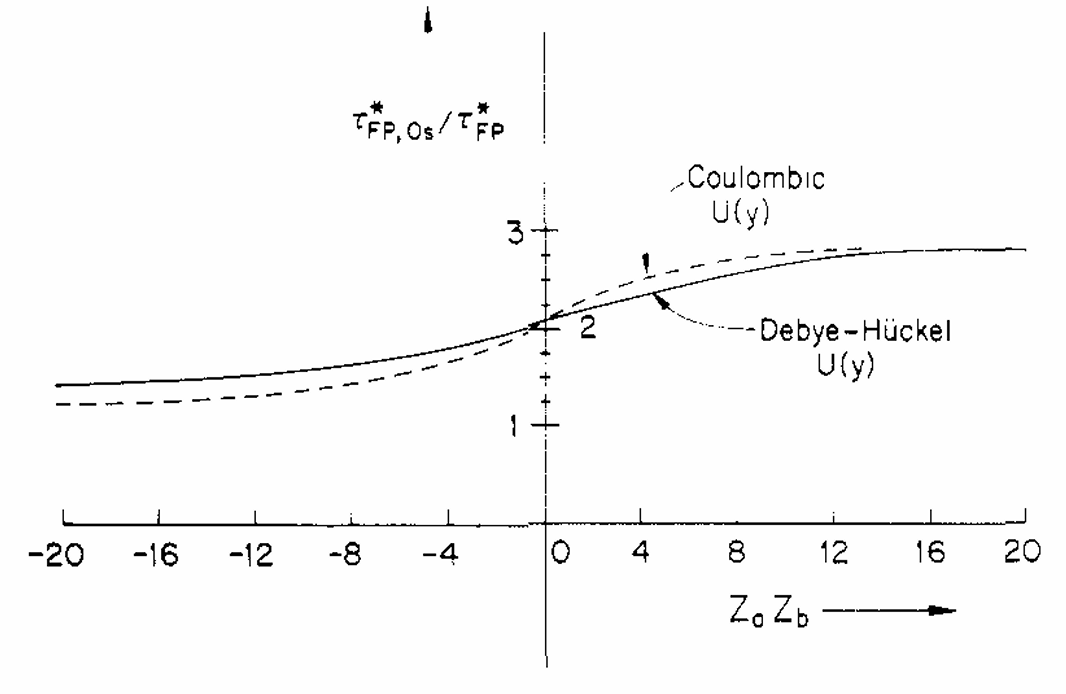
|
|
ABSTRACT: Dynamic effects of a model pair correlation function and Oseen's tensor hydrodynamic interactions are included in the study of the kinetics of protein domain coalescence using the numerical approach of Zientara, Nagy, and Freed. Modifications to the previously reported results due to hydrodynamic drag and either Debye-Hückel or Coulombic electrostatic forces are presented. A model domain pair correlation function is also incorporated to more accurately simulate the spatial and energetic aspects of hydrophobic bonding. Applying this extended model, the variation of coalescence lifetimes with ionic strength and temperature is then calculated and discussed with reference to published experimental data. A frequently observed but anomalous temperature variation in a renaturation rate constant is explained by our results.
|
|
|
A Cryostat for Investigating Spin Polarized Hydrogen
B. Yurke, J.S. Denker, B.R. Johnson, J.H. Freed, and D.M. Lee.
Physica B 107, 521-522 (1981).
<doi: 10.1016/0378-4363(81)90563-5>
Publication #102
|
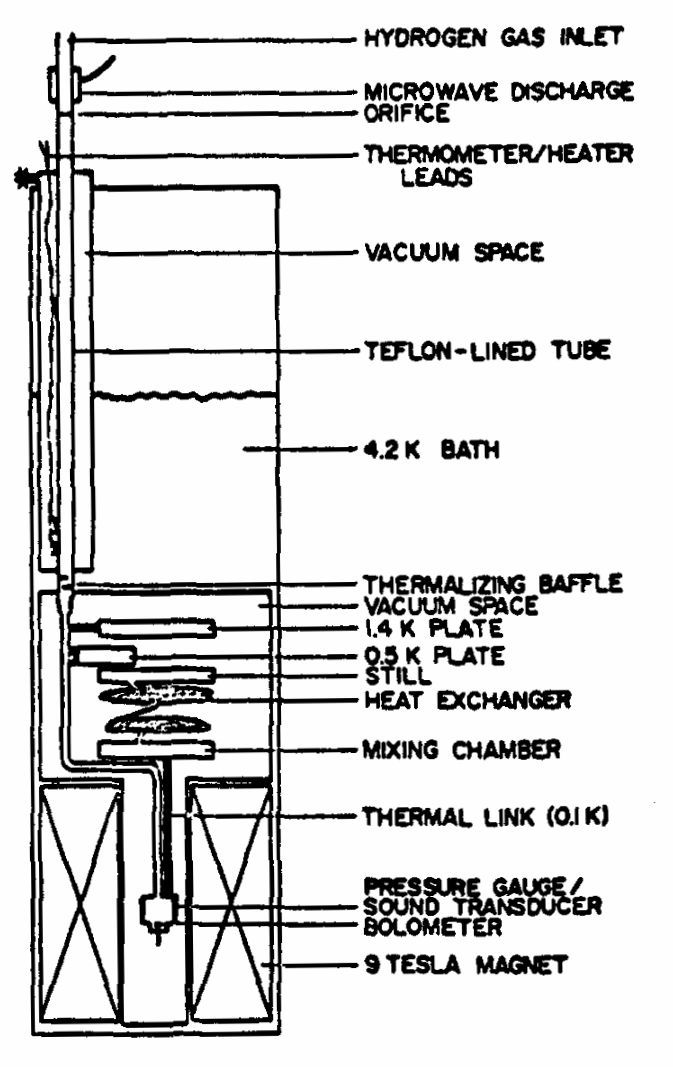
|
|
ABSTRACT: A low temperature spin polarized hydrogen apparatus is discussed. Fluxes into the hydrogen cell of 1014 atoms/sec have been achieved. Hydrogen atom concentrations of 1 × 1016 atoms∕cm3 have been observed in fields of 9 Tesla and at 0.6 K, as determined by strain gauge measurements and calorimetric methods.
|
|
|
UHV ESR and CREMSEE: Two Novel Surface Techniques
M. Nilges and J.H. Freed.
Chem. Phys. Lett. 82, 203-207 (1981).
<doi: 10.1016/0009-2614(81)85139-1>
Publication #101
|
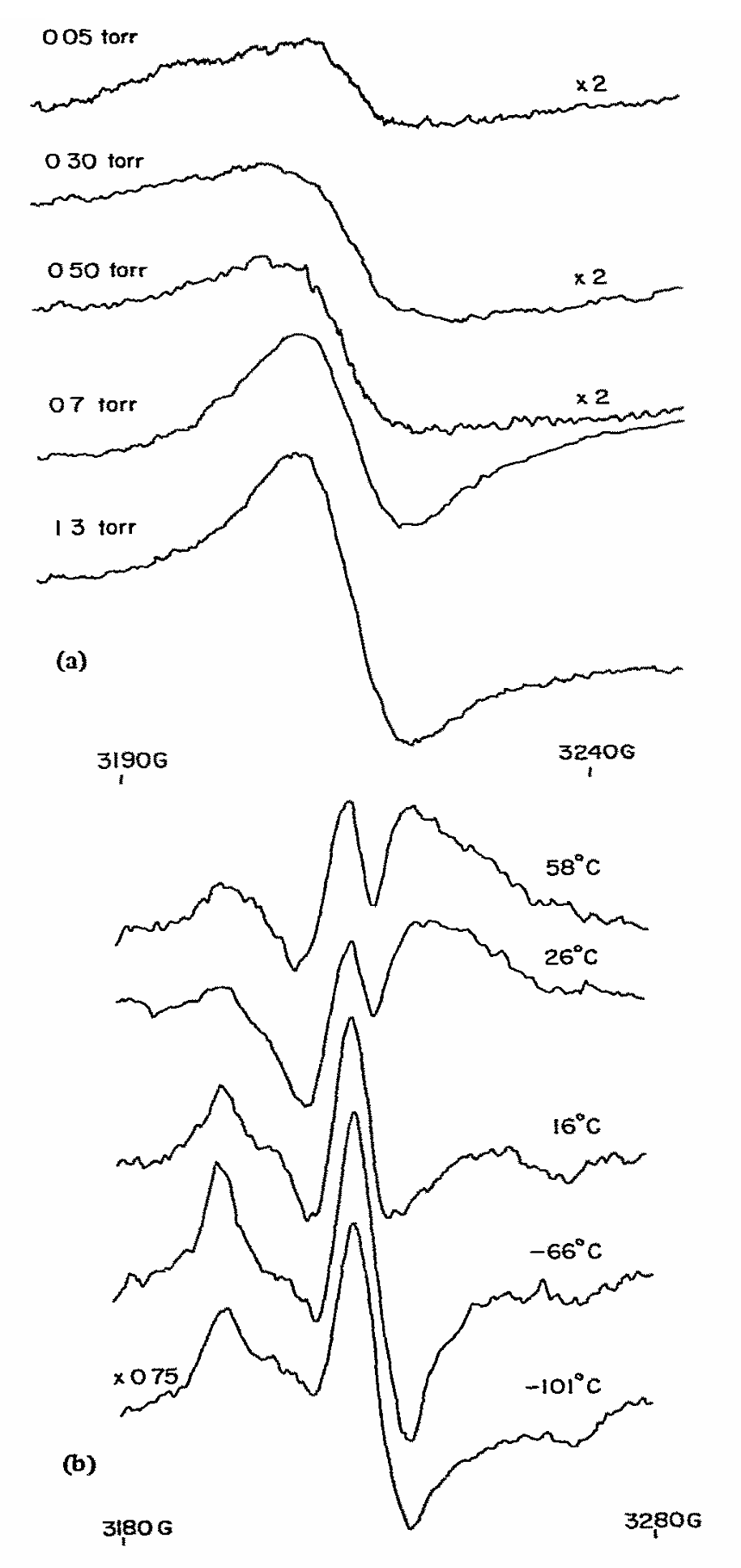
|
|
ABSTRACT: The complementary surface techniques of ultra-high vacuum (UHV) ESR and CREMSEE (cyclotron resonance from microwave-induced secondary electron emission) are described. It is shown in a study of a stable paramagnetic molecule on Cu and Ag surfaces, that it appears to lose its paramagnetism while forming a chemisorbed complex with the metal surface. Control experiments on air-oxidized Cu are also presented.
|
|
|
ESR Studies of NO2 Adsorbed on Surfaces. Analysis of Motional Dynamics
M. Shiotani and J.H. Freed.
J. Phys. Chem. 85, 3873-3883 (1981).
<doi: 10.1021/j150625a033>
Publication #100
|
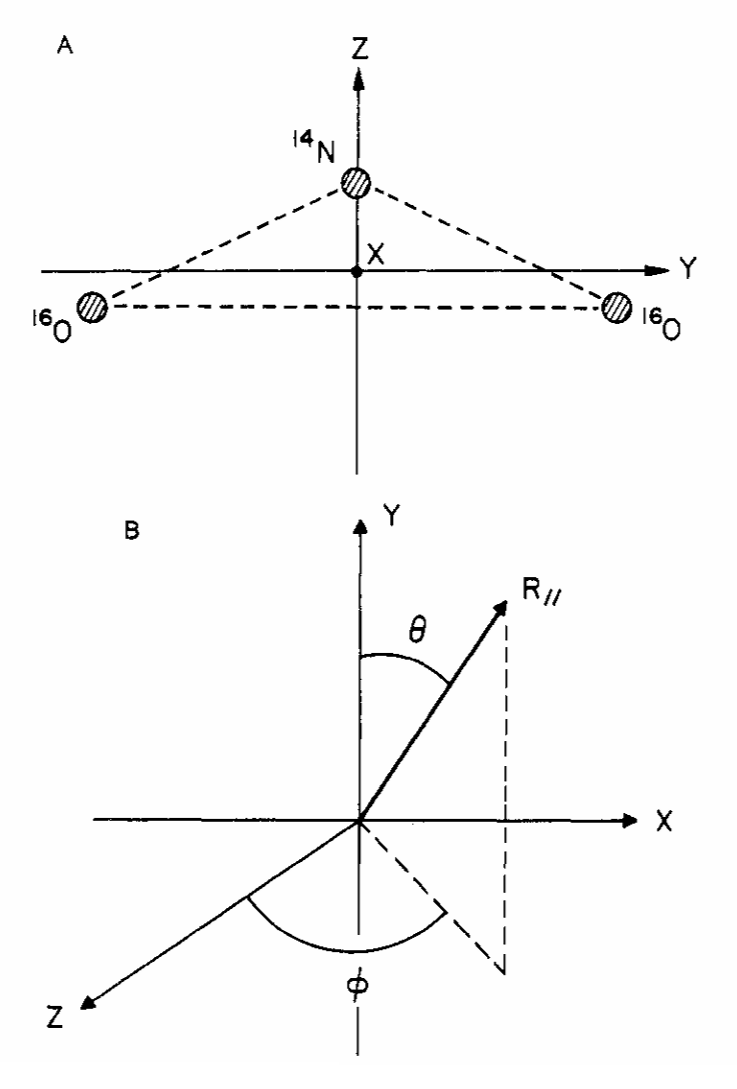
|
ABSTRACT: ESR spectra of NO 2 adsorbed on crushed Vycor glass and on a zeolite were observed in the temperature range 4.8-221 K. Based upon spectral simulations using the slow-motional ESR theory for different models of rotational diffusion, we found that for NO 2 motion on the Vycor surface the following occurs: (1) At 4.8 K, NO 2 still shows residual motional effects. (2) Below 77 K the rotational motion of NO 2 is axially symmetric about the y axis (i.e. parallel to the O-O internuclear axis) such that N ≡ R∥ ∕ R⊥ ≈ 10. The parallel component of the diffusion tensor, R∥, increases with temperature, e.g., R∥ = 5 × 10 5 and 1 × 10 7 s −1 at 4.8 and 77 K, respectively (based on a Brownian diffusion model). (3) Above 77 K, R⊥ increases with temperature so that almost isotropic rotational diffusion is estimated at 185 K (i.e., R∥ = 1 × 107 s−1 and N = 1.3), presumably due to a translational diffusion mechanism. (4) There are discrepancies between the observed and calculated line shapes especially for T ≲ 77 K. None of the models used could improve the agreement, and possible improvements of the models are outlined. A quantum mechanical analysis of the 4.8 K spectrum implies either a renormalized moment of inertia for NO2 that is orders of magnitude greater than that in the gas phase and∕or a strong orientation-dependent interaction potential with the surface. In either case it is suggested that the y axis should be parallel to the surface. Additional line broadening probably due to an exchange process modulated by translational diffusion rapidly increases with temperature above 77 K, even though the NO2 surface concentration decreases with increasing temperature. Distinct differences in motional dynamics are observed for NO2 on the zeolite support. |
|
|
A UHV-ESR Study of the Reaction of NO2 Adsorbed on Copper
M. Nilges, G. Barkley, M. Shiotani, and J.H. Freed.
J. de Chim. Phys. 78, 909-919 (1981).
Publication #99
|
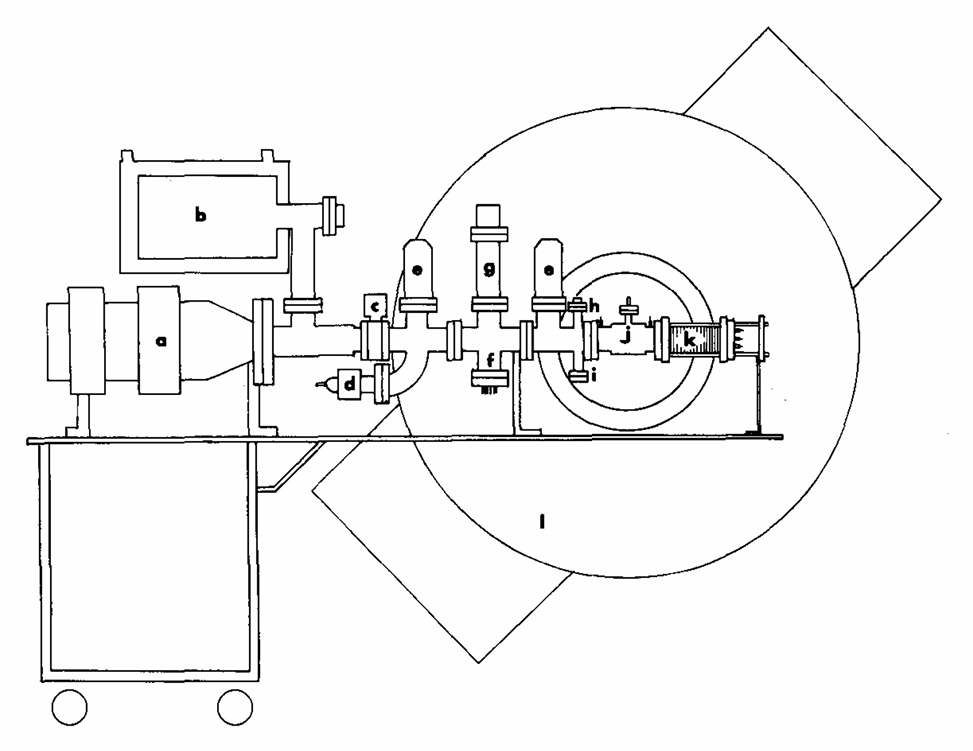
|
|
ABSTRACT: We describe in detail an experimental design which permits the high sensitivity study of ESR from paramagnetic species that may form on clean metal (and oxide) surfaces. The heart of the design is the ultra-high vacuum ESR cavity. The new phenomenon CREMSEE (cyclotron resonance from microwave–induced secondary electron emission), which is observed under high vacuum conditions, is characterized in some detail. In particular, it is found to be a sensitive indicator of surface bonding. The results of a detailed study of the reaction of NO2 with Cu performed in the UHV-ESR system are presented. It is shown that the initial oxidation process may be monitored by CREMSEE, while an ESR signal is only seen when H2O is present. The primary role of the H2O is to form a hydrated surface copper complex which is magnetically concentrated but microcrystalline. These results are then compared with a study on supported Cu performed under conventional vacuum conditions.
|
|
|
Classical Time-Correlation Functions and the Lanczos Algorithm
G. Moro and J.H. Freed.
J. Chem. Phys. 75, 3157-3159 (1981).
<doi: 10.1063/1.442375>
Publication #98
|
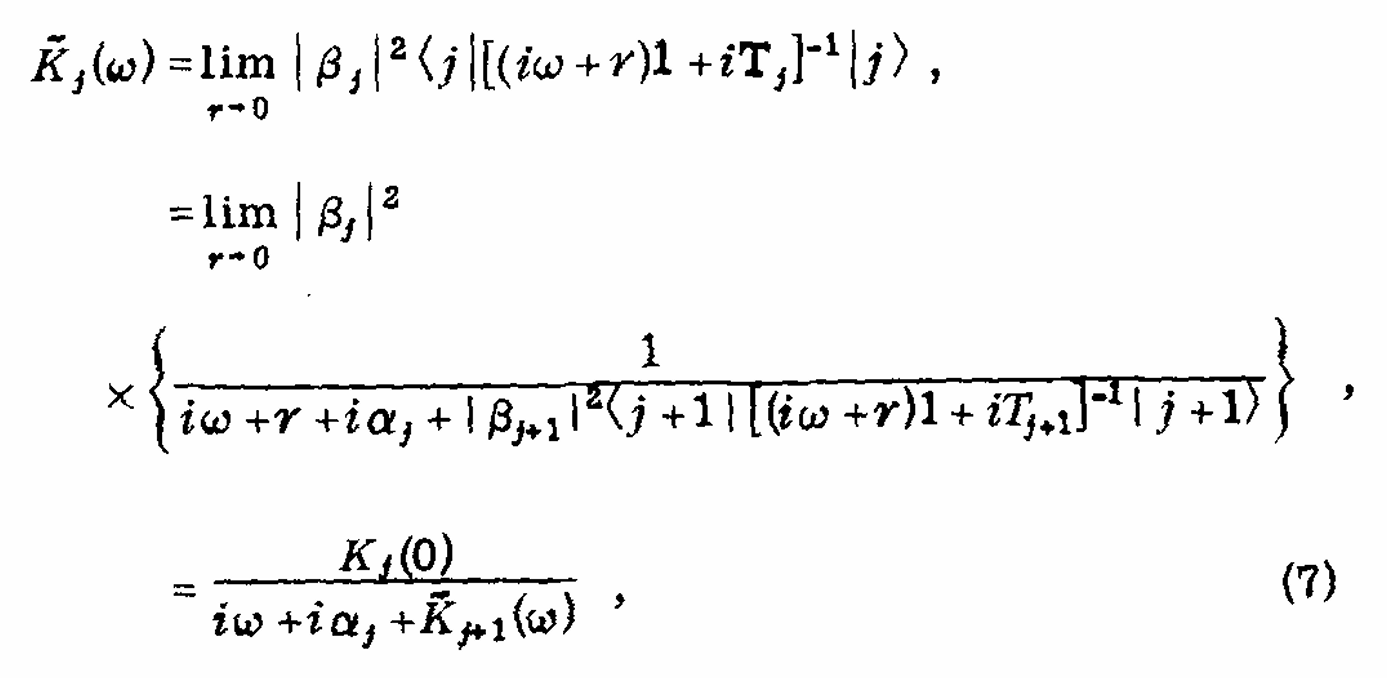
|
|
ABSTRACT: It is shown how the Lanczos algorithm (LA) can be applied to classical systems, leading naturally to a continued fraction representation equivalent to that obtained by Mori for the correlation functions. The relation of the LA to the method of moments suggests its utility for study of the theoretical structure, while the computational efficiency of the LA suggests its use for computer calculation of correlation functions. (AIP)
|
|
|
Rotational Jumps of the Tyrosine Side Chain in Crystalline Enkephalin. 2H NMR Line Shapes for Aromatic Ring Motion in Solids
D.M. Rice, R.J. Wittebort, R.G. Griffin, E. Meirovitch, E.R. Stimson, Y.C. Meinwald, J.H. Freed, and H.A. Scheraga.
J. Am. Chem. Soc. 103, 7707-7710 (1981).
<doi: 10.1021/ja00416a002>
Publication #97
|
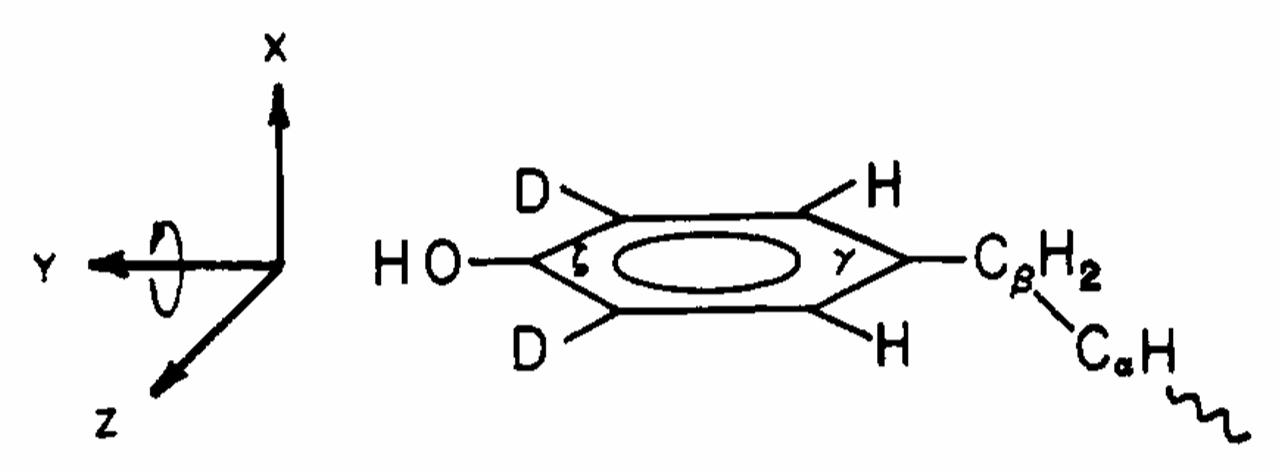
|
|
ABSTRACT: Deuterium NMR spectra of polycrystalline [tyrosine-3,5-2H2] [Leu5]enkephalin show that the aromatic tyrosyl ring of this pentapeptide is executing 180° flips about the Cβ-Cγ axis in the solid state. Specifically, the axially symmetric powder pattern observed at low temperature collapses to an axially asymmetric pattern with η ≃ 0.6 at high temperature. Computer simulations of the NMR line shapes, which account for spectral distortions induced by the quadrupole echo technique, indicate that at room temperature the flipping rate is approximately 5 × 104 s−1 and that it increases to about 106 s−1 at 101 °C.
|
|
|
Effects of Defect Annealing on Concentration-Dependent ESR Spectra from Hydrated Dimyristoylphosphatidylcholine
E. Meirovitch and J.H. Freed.
J. Phys. Chem. 85, 1617-1619 (1981).
<doi: 10.1021/j150612a001>
Publication #96
|
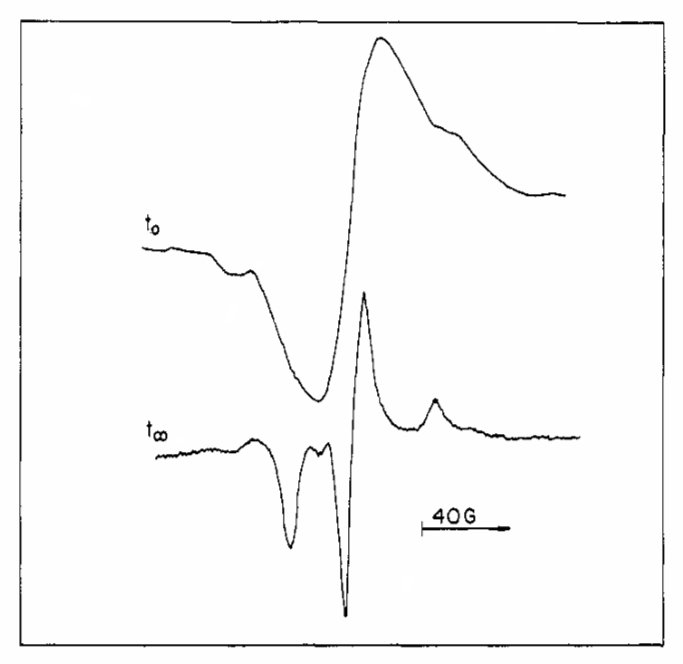
|
|
ABSTRACT: This work reports the dramatic ESR spectral changes which occur for fully hydrated multilayers of dimyristoylphosphatidylcholine (DMPC) having significant concentrations of spin-label, which have been allowed to anneal over a period of 30 days. The ESR spectra initially show concentration-dependent line broadening but then gradually become characteristic of concentration-independent spectra. Spin concentration measurements show no significant loss of radical concentration. These observations may be related to recent work of Chan and Webb showing that intially there are extensive structural defects, which gradually anneal out in the same time period.
|
|
|
Calculation of ESR Spectra and Related Fokker-Planck Forms by the use of the Lanczos Algorithm
G. Moro and J.H. Freed.
J. Chem. Phys. 74, 3757-3773 (1981).
<doi: 10.1063/1.441604>
Publication #95
|
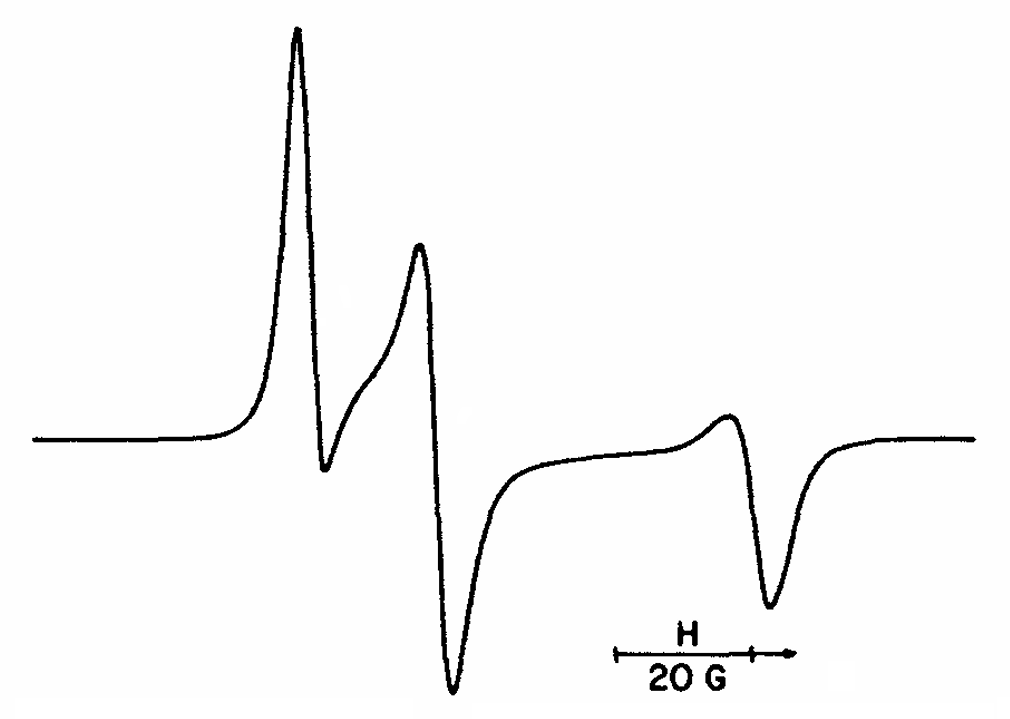
|
|
ABSTRACT: The applicability of the Lanczos algorithm in the general ESR (and NMR) line shape problem is investigated in detail. This algorithm is generalized to permit tridiagonalization of complex symmetric matrices characteristic of this problem. It is found to yield very accurate numerical solutions with at least order of magnitude reductions in computation time compared to previous methods. It is shown that this great efficiency is a function of the sparsity of the matrix structure in these problems as well as the efficiency of selecting an approximation to the optimal basis set for representing the line shape problem as distinct from actually solving for the eigenvalues. Furthermore, it is found to aid in the analysis of truncation to minimize the basis set (MTS), which becomes nontrivial in complex problems, although the efficiency of the method is not very strongly dependent upon the MTS. It is also found that typical Fokker–Planck equations arising from stochastic modeling of molecular dynamics have the property of being representable by complex–symmetric matrices that are very sparse, so calculation of associated correlation functions can be very effectively implemented by the Lanczos algorithm. It is pointed out that large problems leading to matrices of very large dimension can be efficiently handled by the Lanczos algorithm.
|
|
|
ESR Studies of O2- Adsorbed on Ti Supported Surfaces: Analysis of Motional Dynamics
M. Shiotani, G. Moro, and J.H. Freed.
J. Chem. Phys. 74, 2616-2640 (1981).
<doi: 10.1063/1.441334>
Publication #94
|
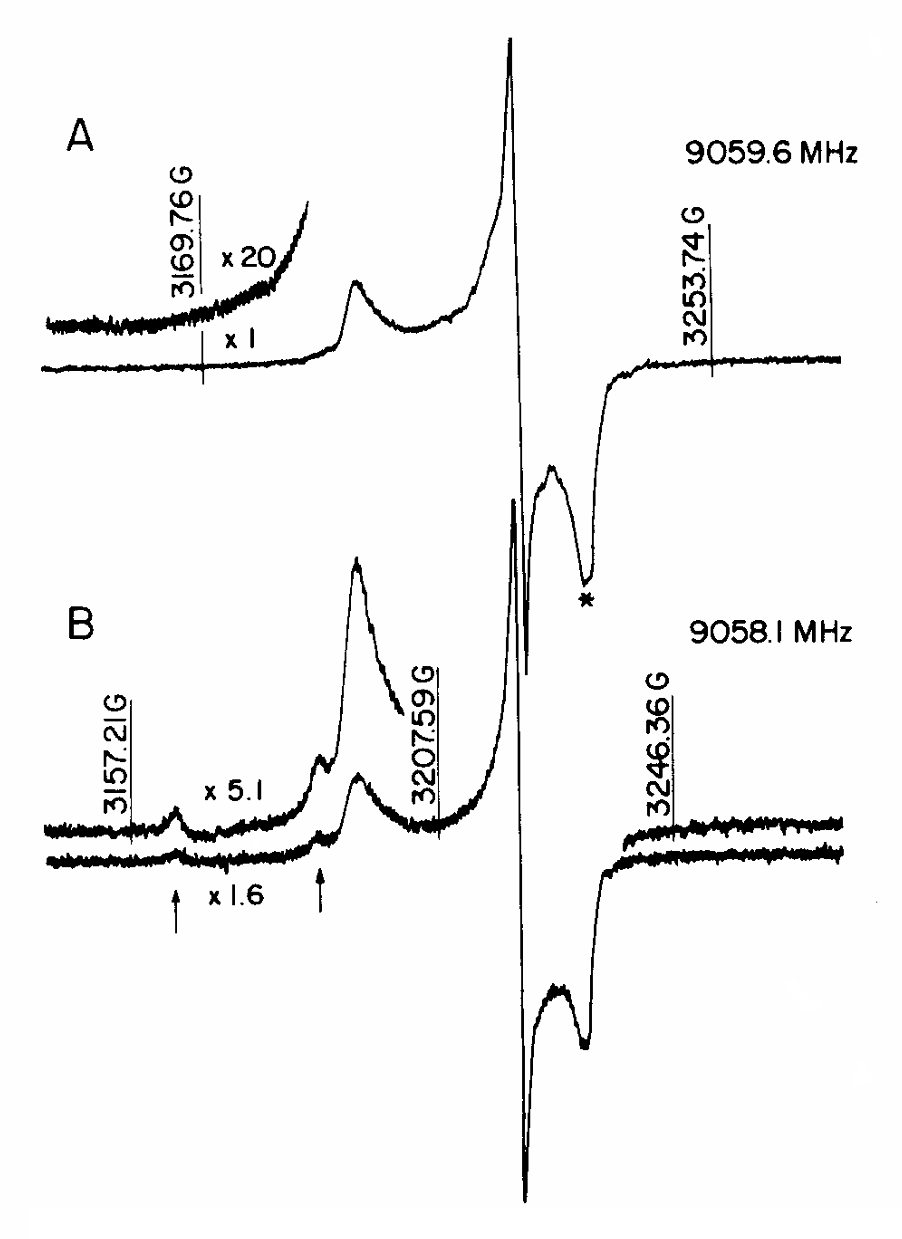
|
|
ABSTRACT: Temperature-dependent ESR spectra of O−2 adsorbed on Ti ions supported on porous Vycor glass were observed over the range 4.2 to 400 °K. These spectra were obtained under normal high vacuum conditions as well as under UHV conditions (P≤10−9 Torr) and are very well resolved. It was observed that the line position of the g̃ tensor component that is perpendicular to the internuclear axis of O−2 remained constant with temperature, whereas the other two components of the g̃ tensor shift in position with temperature, and are accompanied by drastic line shape changes. This observation indicates that the molecular motion of O−2 on the surface is highly anisotropic, consisting essentially of planar rotation about the axis perpendicular to the internuclear axis of O−2 and parallel to the normal to the surface. Furthermore, the observation of nonequivalent 17O hfs of O−2 suggests that the internuclear axis of O−2 might be tilted slightly from the surface and∕or one oxygen is closer to the Ti4+. The ESR line shapes were simulated for the different possible models: Brownian diffusion, jump diffusion (from weak jump to strong jump), approximate free diffusion, and discrete jump. It was found that the theoretical spectra calculated using the model of weak jump rotational diffusion best fit the observed spectra in the temperature range below 57.4 °K. However, in the temperature range above 57.4 °K, although the Brownian diffusion model seems the best among the models used, none of the present models used could successfully reproduce the observed line shapes. The rotational correlation time τR∥ was found to range between 10−5 sec (below 14.5 °K) and 10−9 sec (263 °K). The values of τR∥ depend strongly on the model used in the lower temperature range, but were essentially independent of model above 100 °K. The activation energy for rotational diffusion was estimated to be 0.5 kcal∕mole above 100 °K. The line shape below 15 °K is independent of temperature, although the O−2 spectrum appears to exhibit residual motional effects. This observation suggests that coherent quantum mechanical motion is predominant below 15 °K. This matter is discussed in some detail, and the appropriate theory to investigate quantum effects on the motional dynamics is outlined including possible isotope effects on the motion. Spectral observation of possible interaction between C2H4 and O−2 on the surface is presented. Also discussed are the techniques for preparing samples with strong well-resolved signals and for removing the other types of O−2 signals, which do not show significant temperature-dependent spectral changes.
|
|
|
Efficient Computation of Magnetic Resonance Spectra and Related Correlation Functions from Stochastic Liouville Equations
G. Moro and J.H. Freed.
J. Phys. Chem. 84, 2837-2840 (1980).
<doi: 10.1021/j100459a001>
Publication #93
|
|
|
ABSTRACT: It is found that a very powerful method for computing slow-motional ESR (and NMR) spectra may be developed by the use of the Lanczos algorithm (LA) modified to tridiagonalize complex-symmetric matrices. It leads to at least order of magnitude reductions in computation time and in computer storage requirements than the commonly used Rutishauser algorithm. This permits the rapid analysis of spectral problems that were previously too difficult. The formal similarity of these problems to those from Fokker-Planck equations in molecular dynamics suggests that the LA should also be a very powerful method for the latter.
|
|
|
|
|
|
|
Diffusion-Controlled Kinetics of Protein Domain Coalescence: Effects of Orientation, Interdomain Forces and Hydration
G.P. Zientara, J.A. Nagy, and J.H. Freed.
J. Chem. Phys. 73, 5092-5106 (1980).
<doi: 10.1063/1.439987>
Publication #91
|
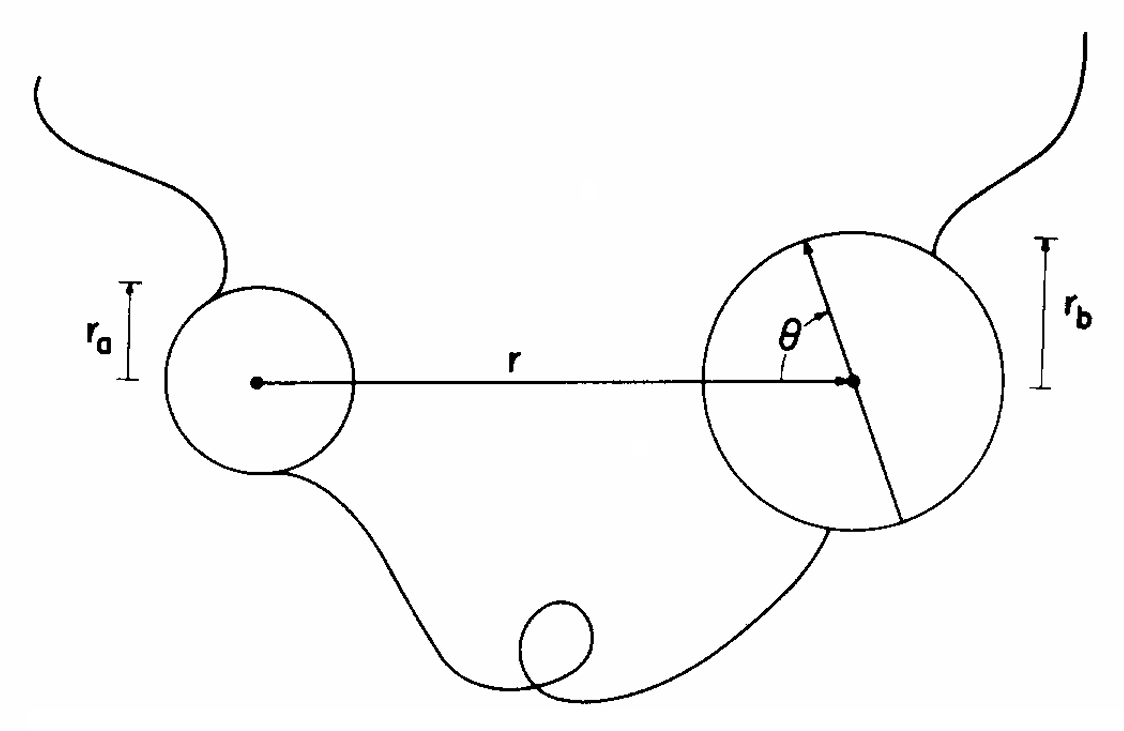
|
|
ABSTRACT: The numerical methods of Zientara and Freed have been used to study the diffusion-controlled kinetics of domain coalescence in order to ultimately consider the time evolution of protein folding. The mean coalescence lifetime for a domain pair has been calculated using the eigenfunction expansion method and finite differences in the solution of a modified form of the Smoluchowski equation. This approach allows for an orientational preference in the domain coalescence process, which has been studied throughout the complete range of reactivities. Three cases of interdomain forces were investigated, one of which represents the shielding of charged domains by ions in solution. As the forces vary from the strongly repulsive to the strongly attractive in these cases, the coalescence lifetime decreases by several orders of magnitude. In addition, a hydration shell model, which provides an activation barrier to coalescence, has been analyzed. The lifetimes resulting from this model were observed to depend strongly on the location and extent of the hydration shell. Anisotropic initial conditions were also incorporated causing, in some cases, non-negligible effects upon the coalescence lifetimes.
|
|
|
Spin-Aligned Hydrogen: Some Considerations for ESR vs. NMR Experiments and Preliminary Observations of H↑ at Low Temperatures
B. Yurke, D. Igner, E. Smith, B. Johnson, J. Denker, C. Hammel, D. Lee, and J.H. Freed.
J. de Phys. 41, C7:177-184 (1980).
Publication #90
|
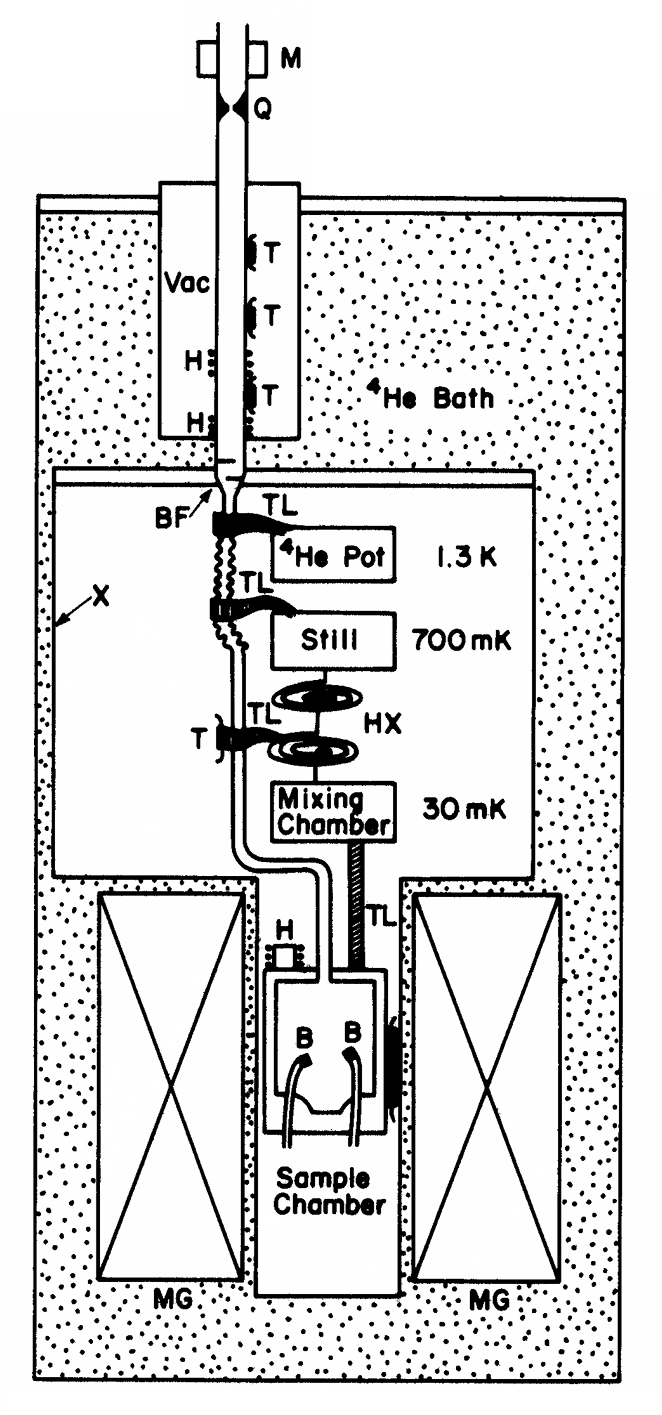
|
|
ABSTRACT: We consider aspects of theoretical sensitivities, spin-relaxation, and radiation damping in proposed low temperature ESR vs. NMR studies on spin-aligned hydrogen (H↑). Also considered are experimental design features. Results are described in a preliminary report of experiments on H↑ at 0.l-0.5°K at 60 kG using bolometer detection. They are consistent with the Silvera-Walraven experiment: viz they imply that H↑ may be stabilized under these conditions when a surface coating of 4He is utilized.
|
|
|
|
|
ESR Studies of Low Water Content 1,2-Dipalmitoyl-sn-Glycero-3-Phosphocholine in Oriented Multilayers. 2. Evidence for Magnetic-Field-Induced Reorientation of the Polar Headgroups
E. Meirovitch and J.H. Freed.
J. Phys. Chem. 84, 3295-3303 (1980).
<doi: 10.1021/j100461a032>
Publication #88
|
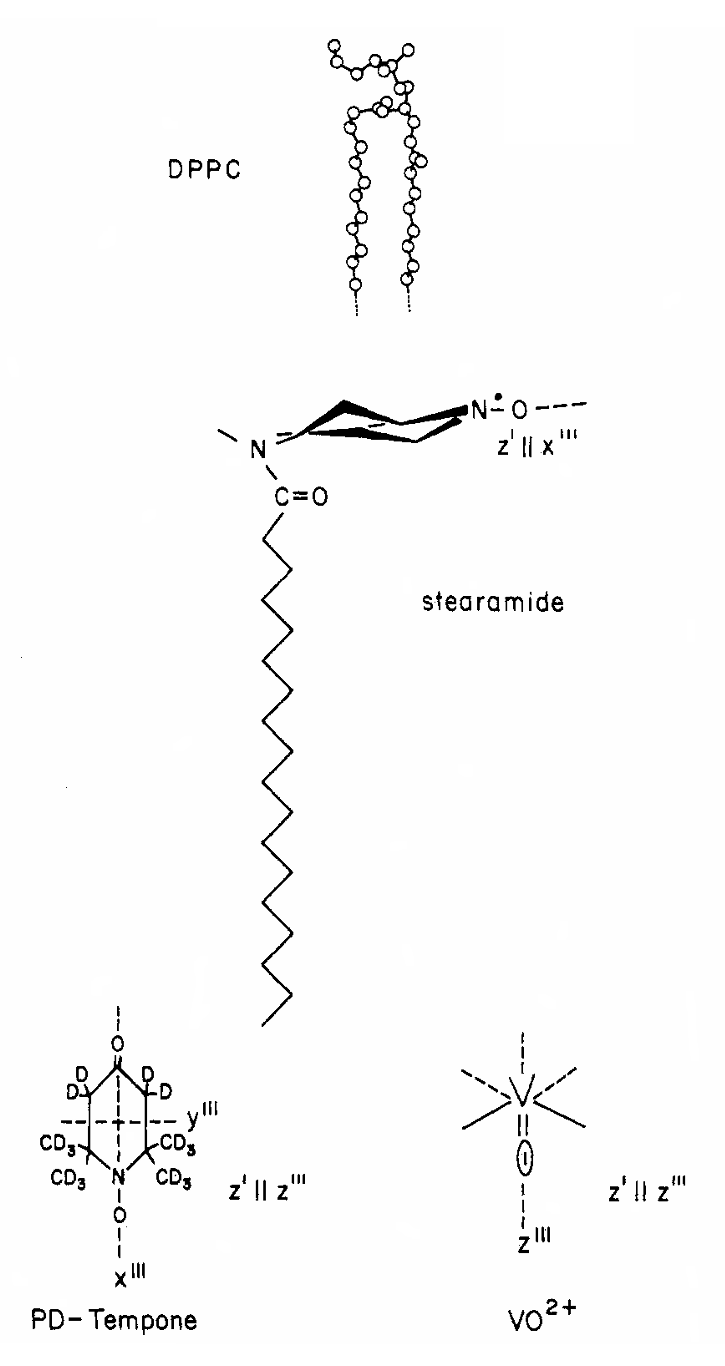
|
|
ABSTRACT: In this continuation of ESR studies on a variety of spin probes incorporated into low-water-content 1,2-di-palmitoyl-sn-glycero-3-phosphocholine (DPPC) bilayers, results with the headgroup-region probes, PD-Tempone and VO2+, are reported for the biaxial phase. The fluidity of the weakly ordered PD-Tempone probe is quite high for higher water content (14 wt %) but decreases for lower water content (9 wt %) (i.e., R⊥, the diffusion coefficient, decreases from 109 to 108 s−1 and shows properties probably related to slowly relaxing local structure in the latter case). The VO2+ ions are, however, immobilized on the ESR time scale and weakly oriented. Both probes show distributions in local director, which, however, become better aligned as H͐, the magnetic field, is tilted into the bilayer plane. Also both probes then have a preferential alignment perpendicular to the projection of H͐ in this plane. These results are taken as definite evidence for cooperative headgroup alignment by the magnetic field. While positive (vs. negative) diamagnetic susceptibility is expected they could not be distinguished by these experiments. Anomalous results from a stearamide probe in the Lα(1) phase, viz. an unusually large observed splitting of 19.05 G with plate samples but typical splitting of 16.1 G with tube samples, is taken as evidence for significant differences in microscopic ordering properties of the DPPC samples resulting from different anchoring constraints imposed by the shapes of the holders. The nature of the biaxiality observed in the biaxial phase by other techniques is considered in the light of these ESR results.
|
|
|
ESR Studies of Low Water Content 1,2-Dipalmitoyl-sn-Glycero-3-Phosphocholine in Oriented Multilayers. 1. Evidence for Long-Range Cooperative Chain Distortions
E. Meirovitch and J.H. Freed.
J. Phys. Chem. 84, 3281-3295 (1980).
<doi: 10.1021/j100461a031>
Publication #87
|
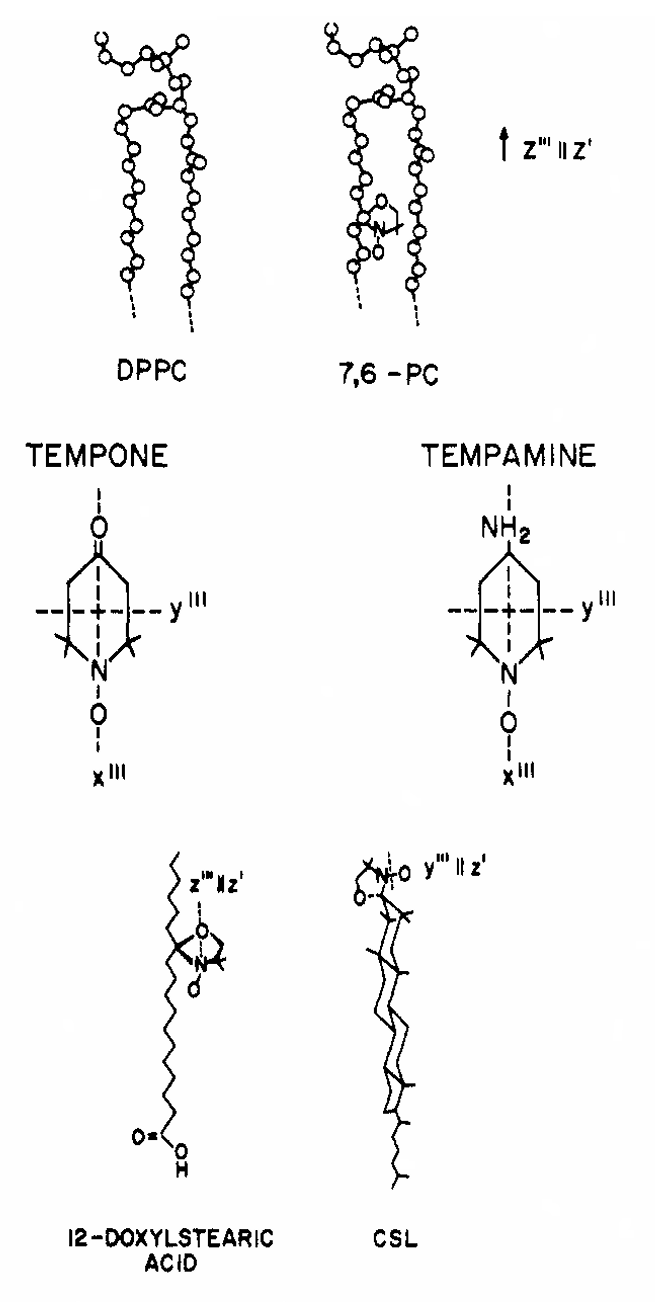
|
|
ABSTRACT: ESR studies on a variety of spin probes incorporated into 1,2-dipalmitoyl-sn-glycero-2-phosphocholine (DPPC) bilayers are reported. Well-oriented and low water content (2-15 wt %) bilayers were prepared by using a recently developed alignment technique. Both the higher temperature lamellar liquid crystalline Lα(1) phase and the lower temperature biaxial gel phase were studied. In the Lα(1) phase there is considerable fluidity, and while all the probes are ordered to varying degrees they undergo fairly rapid rotational reorientation with R̄ (the mean rotational diffusion coefficient) of the order of 0.1 × 109 −l × 109 s and with comparable activation energies of the order of 10 kcal/mol. In this phase, headgroup region probes such as PD-Tempone exhibit spectra as a function of angle θ between the magnetic field and the bilayers that are fully consistent with the well-aligned lamellar morphology. However, the hydrophobic probes such as the spin labeled DPPC (7,6-PC) and cholestane (CSL) exhibit a unique powder-type distribution that appears inconsistent with this lamellar morphology or even with random (or Gaussian) distributions due to sample morphology. On the basis of extensive line-shape analyses (including the use of a new slow-tumbling formulation in which θ may be varied) it is shown that satisfactory fits may be achieved only by using a special distribution corresponding to a coherent two-dimensional distribution of local directors. This is consistent with a cooperative distortion wave persisting for time scales long compared to ESR time scales. In particular, since the hydrophobic probes are expected to report on the chain region of the DPPC bilayers, it is argued that these distortion waves involve bends or kinks of the chains, but do not affect the headgroup region in this Lα(1) phase. The implications of these results are also examined from the point of view of measurements by other techniques. It is adso shown that similar spectra are obtained from the highly ordered smectic Bc phases of thermotropic liquid crystals implying similar cooperativity. In the biaxial phase the 7,6-PC appears to be immobilized on the ESR itime scale, but CSL, which is highly ordered in this phase, is still reorienting at about 109 s. In both cases evidence of the cooperative distortion mode is found in the less well-resolved spectra. Further evidence from headgroup region probes indicates that this region is now affected by this mode. Significant changes in the hyperfine tensor of probes in this phase is taken as evidence of intermolecular interactions. We have also studied samples using cylindrical tubes as holders. These yield macroscopically unoriented morphologies, with characteristic line shapes commonly observed in previous spin-probe and spin-label studies of biological membranes, but usually interpreted in terms of mixtures of mobile and immobile sites, an interpretation brought into question by the detailed analyses of this work. Details and implications of the slow-tumbling line-shape simulations as a function of θ are also discussed.
|
|
|
Electron Spin Resonance Studies of Anisotropic Ordering, Spin Relaxation, and Slow Tumbling in Liquid Crystalline Solvents. 4. Cholestane Motions and Surface Anchoring in Smectics
E. Meirovitch and J.H. Freed.
J. Phys. Chem. 84, 2459-2472 (1980).
<doi: 10.1021/j100456a023>
Publication #86
|
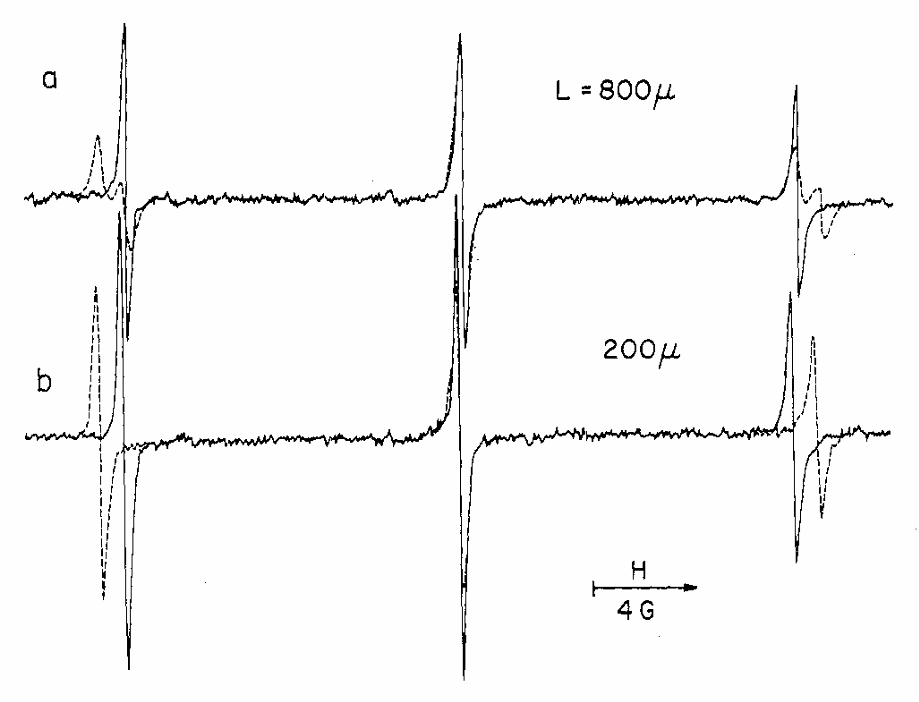
|
|
ABSTRACT: ESR line shape studies of cholestane (CSL) in monodomain liquid crystals of HOAB, 40,6, and 40,8 oriented between glass surfaces, are reported. The line shapes are analyzed in terms of the Polnaszek-Bruno-Freed theory for spectra from slow tumbling probes in ordered fluids. Preparation of monodomain smectic crystals, free from previously observed effects of static director distribution due to magnetic torques, and therefore adequate for accurate studies of dynamic line shapes, both as a function of temperature and orientation, is described. CSL exhibits fast motion in HOAB but slow motion in 40,8 and 40,6. In the nematic phases the anisotropy in the diffusion tensor (N ≈ 5) is consistent with the geometry of the probe molecule, as expected for simple Brownian type of diffusion in a mean potential field. In the smectic phases, an "apparent" anisotropy was found by simulations to be substantially greater than predicted by the molecular configuration and appears to be increasing dramatically as the temperature is decreased. It is argued that this is untenable in the context of the model. Instead, it is shown that an approximate model of fluctuating torques could be successfully used to interpret the apparent anomalies. These observations are interpreted in terms of the effect of the local solvent structure on the nature of the overall dynamic mode of the probe, implying considerable local cooperativity in these smectic phases. It is also shown by preliminary studies with the PD-Tempone probe that the very well-aligned samples may experience substantial elastic distortions with magnetic fields of 3 kG when the field is tilted relative to the normal to the plates. Since this is contrary to predictions of previous theory, a new theoretical analysis is given which suggests that (1) the conditions of weak vs. strong anchoring are different in the nematic vs. smectic phases and (2) it is quite possible to have weak anchoring in the smectic phase of a sample which exhibits rather strong anchoring in the nematic phase.
|
|
|
Slow-Motional ESR Spectra for Vanadyl Complexes and Their Model Dependence
R.F. Campbell and J.H. Freed.
J. Phys. Chem. 84, 2668-2680 (1980).
<doi: 10.1021/j100457a038>
Publication #85
|
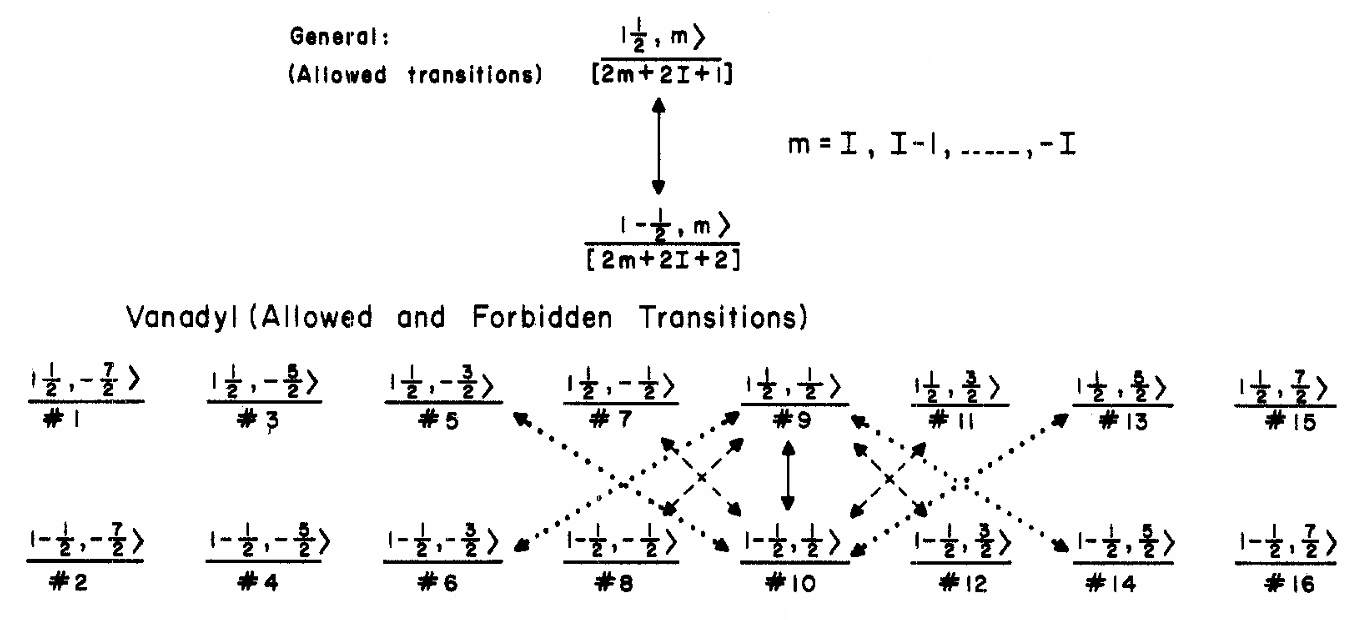
|
|
ABSTRACT: The slow-motional ESR analysis appropriate for vanadyl(IV) ions is developed with an accurate treatment of the nonsecular contributions. This results in good agreement between theory and experiments on VO(acac2(pm)) in toluene with axially symmetric magnetic parameters over the whole motional range when a Brownian motion model is used. It is also found that a slightly modified motional narrowing theory based upon the stochastic Liouville equation leads to improved agreement between theory and experiment for VO(acac)2 in toluene which also obeys a Brownian motion model. It is shown that vanadyl slow-tumbling spectra are very sensitive to model (even more so than nitroxides). In particular, experiments on VO(H2O)52+ in aqueous solution are approximately fit by a model of moderate jump, while those on VO(NCS)42− could only be crudely fit by temperature-dependent variations in model. The theory given here is appropriate for any S = 1∕2 radical with a single nuclear spin I provided only the high-field approximation is valid, so that nonsecular terms may be treated by a perturbation approach. The effects of (1) angular-dependent transition probabilities and (2) field- vs. frequency-swept spectra upon the slow-motional theory are also discussed.
|
|
|
Spin-Echoes for Diffusion in Bounded, Heterogeneous Media: A Numerical Study
G.P. Zientara and J.H. Freed.
J. Chem. Phys. 72, 1285-1292 (1980).
<doi: 10.1063/1.439190>
Publication #84
|
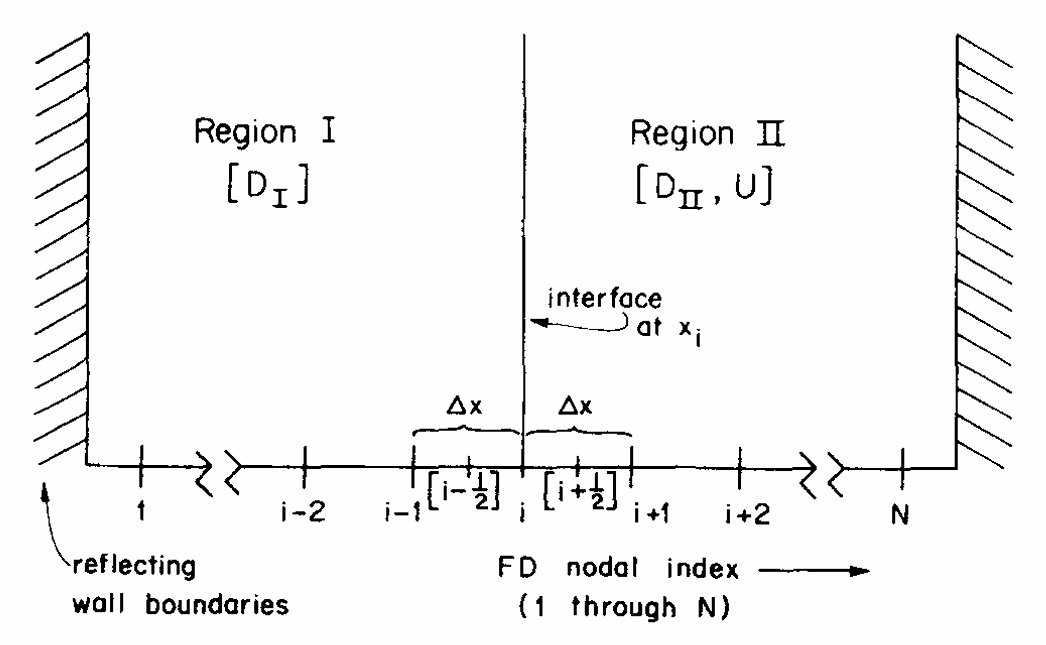
|
|
ABSTRACT: The diffusive behavior of spin-bearing species in a bounded heterogeneous medium is analyzed in a manner appropriate for spin echo experiments in the presence of field gradients. A numerical method based upon the stochastic Liouville equation (SLE) is discussed that includes the discontinuities in transport and solubility properties due to the different spatial regions. The double step computational algorithm, which takes advantage of the different time scales of diffusive and spin-quantum phenomena, is then introduced as a general approximate solution of the time dependent SLE. This method is applied to the calculation of the decay of spin echo amplitudes, and it suggests a new approach for analyzing such experiments in terms of the microscopic details and chemical properties of heterogeneous systems.
|
|
|
Theory of Chemically Induced Dynamic Spin Polarization. 5. Orientation-Dependent Effects
G.P. Zientara and J.H. Freed.
J. Phys. Chem. 83, 3333-3344 (1979).
<doi: 10.1021/j100489a006>
Publication #83
|
|
|
ABSTRACT: The Pedersen-Freed theory for chemically induced dynamic spin polarization CIDN(E)P is generalized to include the effects of anisotropic reactivities and anisotropic exchange interactions on the radical-pair mechanism. Detailed results are given for the simple case in which only one radical exhibits anisotropy that is approximated by a cosine distribution, and the rotational and translational motions are described by Brownian diffusion models. The primary effect upon CIDNP is the reduction in Λ, the reaction probability for the full collision. This effect can be rather accurately approximated by the use of an "effective" spherically symmetric specific rate constant, which depends, to some extent, on the rotational diffusion coefficient due to the effect of rotational relaxation. Thus CIDNP effects are rather well approximated by a spherically symmetric theory with a renormalized Λ. In the absence of reactivity, CIDEP effects for our model are reasonably well approximated by a spherically symmetric theory with a renormalized exchange interaction, especially for the asymptotic polarizations for large exchange interactions. When, however, there are orientation-dependent reactivities present with substantially greater orientation dependence than the exchange interaction, then significant deviations from a spherically symmetric theory are predicted even for asymptotic polarizations. The relationship to recent experiments is briefly discussed in this light. Anisotropic initial conditions are typically found to be relaxed by the effective rotational diffusion before they can substantially affect the CIDN(E)P observables.
|
|
|
Stochastic Modeling of Generalized Fokker-Planck Equations. I.
A.E. Stillman and J.H. Freed.
J. Chem. Phys. 72, 550-566 (1980).
<doi: 10.1063/1.438942>
Publication #82
|
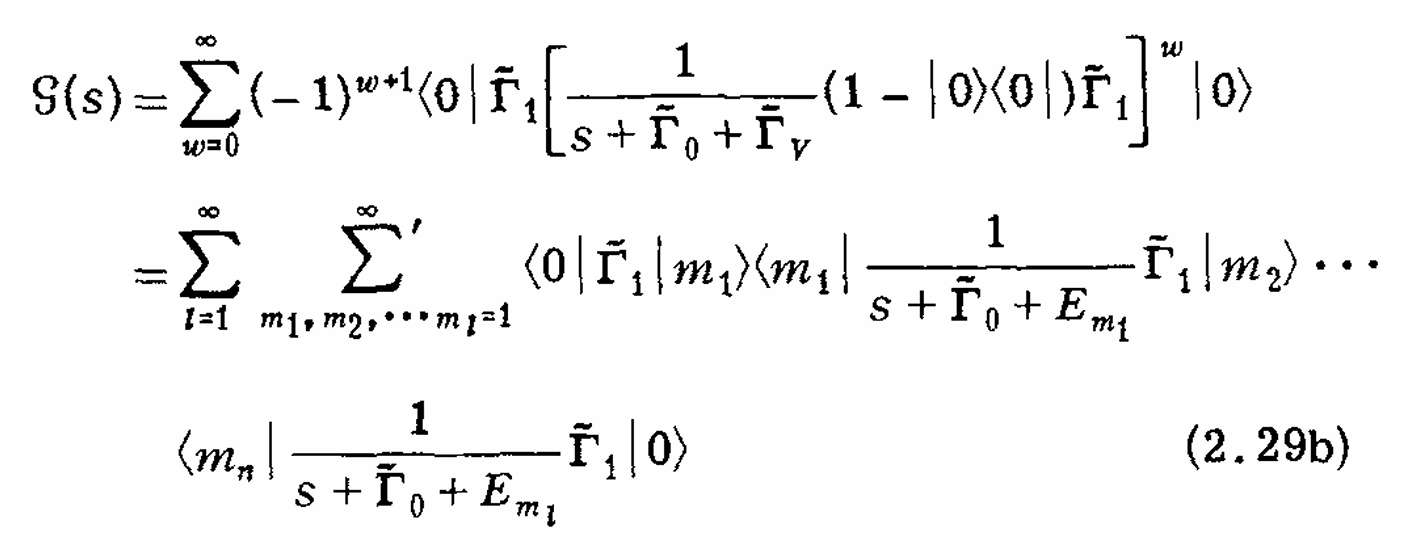
|
|
ABSTRACT: A relatively simple method is developed whereby the many-body features of a typical generalized Fokker–Planck equation (GFPE) for a diffusing molecule are first replaced by stochastic bath variables that are assumed to be Markovian. Then the combined molecular and bath variables are characterized as a multidimensional Markov process obeying a stochastic–Liouville equation, which is, in general, incomplete, because it ignores the back reaction of the molecule on the bath variables. In the final step, the equation is completed by subjecting it to the appropriate constraints required for detailed balance. In this form the augmented Fokker–Planck equation (AFPE) properly describes relaxation to thermal equilibrium, and, for the appropriate limiting conditions, it reduces to the classical Fokker–Planck equation. This procedure for stochastic modeling of GFPE is both an improvement on and a generalization of a method previously outlined by Hwang, Mason, Hwang, and Freed (HMHF). Detailed illustrations of AFPE's are presented for the simple case of a planar rotator subjected to fluctuating torques, and these models are extended to the case of three-dimensional rotational diffusion. Examples include fluctuating torque models related to that used by HMHF. It is shown that only if the fluctuating torque is independent of the orientation of the molecule (more precisely of any fluctuating equilibrium orientation of the molecule), does the model become equivalent to the usual generalized Langevin equation. Otherwise, more general nonlinear models are obtained, which, however, are easily handled by the present methods. Models related to the slowly relaxing local structure (SRLS) model of Polnaszek and Freed are also developed. They are shown to be a consequence of requiring relaxation to the instantaneous value of the fluctuating potential associated with the torque, whereas the fluctuating torque models are a consequence of requiring relaxation to a uniform orientational distribution. They differ further in that the SRLS models are "nonfrictional". For these reasons we characterize the fluctuating torques as being "collision-induced" and the SRLS as being "structure-induced".
|
|
|
The Variational Method and the Stochastic-Liouville Equation: III. Infinite Elements for CIDN(E)P
G.P. Zientara and J.H. Freed.
J. Chem. Phys. 71, 744-749 (1979).
<doi: 10.1063/1.438361>
Publication #81
|
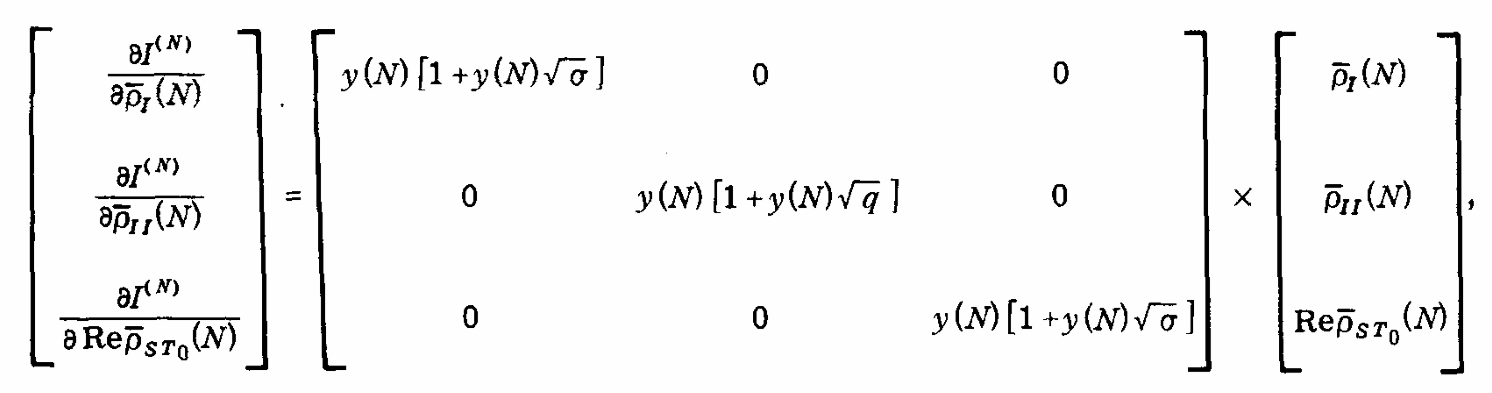
|
|
ABSTRACT: The variational finite-element method introduced by Zientara and Freed for the solution of the stochastic-Liouville equation is modified to utilize the advantages of an infinite outer element. Within this infinite element, the correct asymptotic forms of the solutions may be used, and they may be matched to those of the inner finite elements. Large reductions in the computational effort are realized by this scheme while maintaining high accuracy as illustrated in the example of high-field chemically induced dynamic spin polarization. Applicability of this method extends to solutions of partial differential equations in chemical physics characterized by a large spatial region with simple interactions and a restricted region in which more complex behavior occurs, such as is found in treatments of chemical reactions modulated by liquid state diffusive processes and in scattering theory in quantum mechanics.
|
|
|
Is Spin-Aligned Hydrogen a Bose Gas?
J.H. Freed.
J. Chem. Phys. 72, 1414-1415 (1980).
<doi: 10.1063/1.439224>
Publication #80
|
|
|
ABSTRACT: The Born−Oppenheimer approximation is used to discuss Bose statistics of spin aligned H atoms under conditions of low temperature, high magnetic field and high density. (AIP)
|
|
|
The Variational Method and the Stochastic-Liouville Equation. II. ESR Spectral Simulation Via Finite Elements
A.E. Stillman, G.P. Zientara, and J.H. Freed.
J. Chem. Phys. 71, 113-118 (1979).
<doi: 10.1063/1.438108>
Publication #79
|
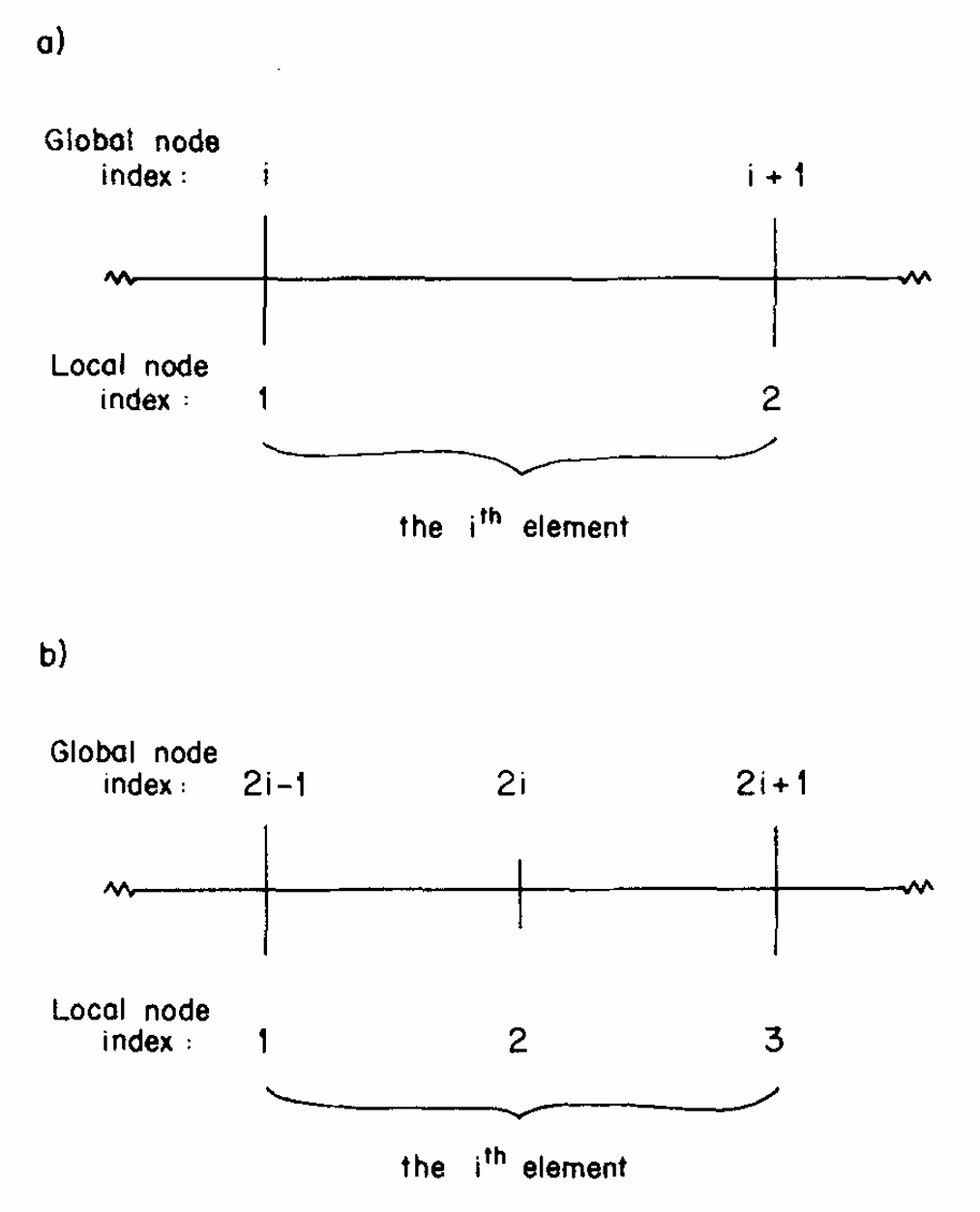
|
|
ABSTRACT: A Galerkin finite element (FE) method, closely related to the variational FE method of Zientara and Freed, is developed for the solution of the stochastic Liouville equation (SLE). The particular illustrative application considered is the ESR spectral simulation of the simple axially symmetric g tensor problem. Both linear and quadratic interpolating functions are considered. It is found for this simple case that the Galerkin FE is almost, but not quite, as efficient as eigenfunction expansions (EE). However, the potential advantages of the Galerkin FE in more complex problems are discussed.
|
|
|
Slow Motional NMR Lineshapes for Very Anisotropic Diffusion: I=1 Nuclei
E. Meirovitch and J.H. Freed.
Chem. Phys. Lett. 64, 311-316 (1979).
<doi: 10.1016/0009-2614(79)80520-5>
Publication #78
|
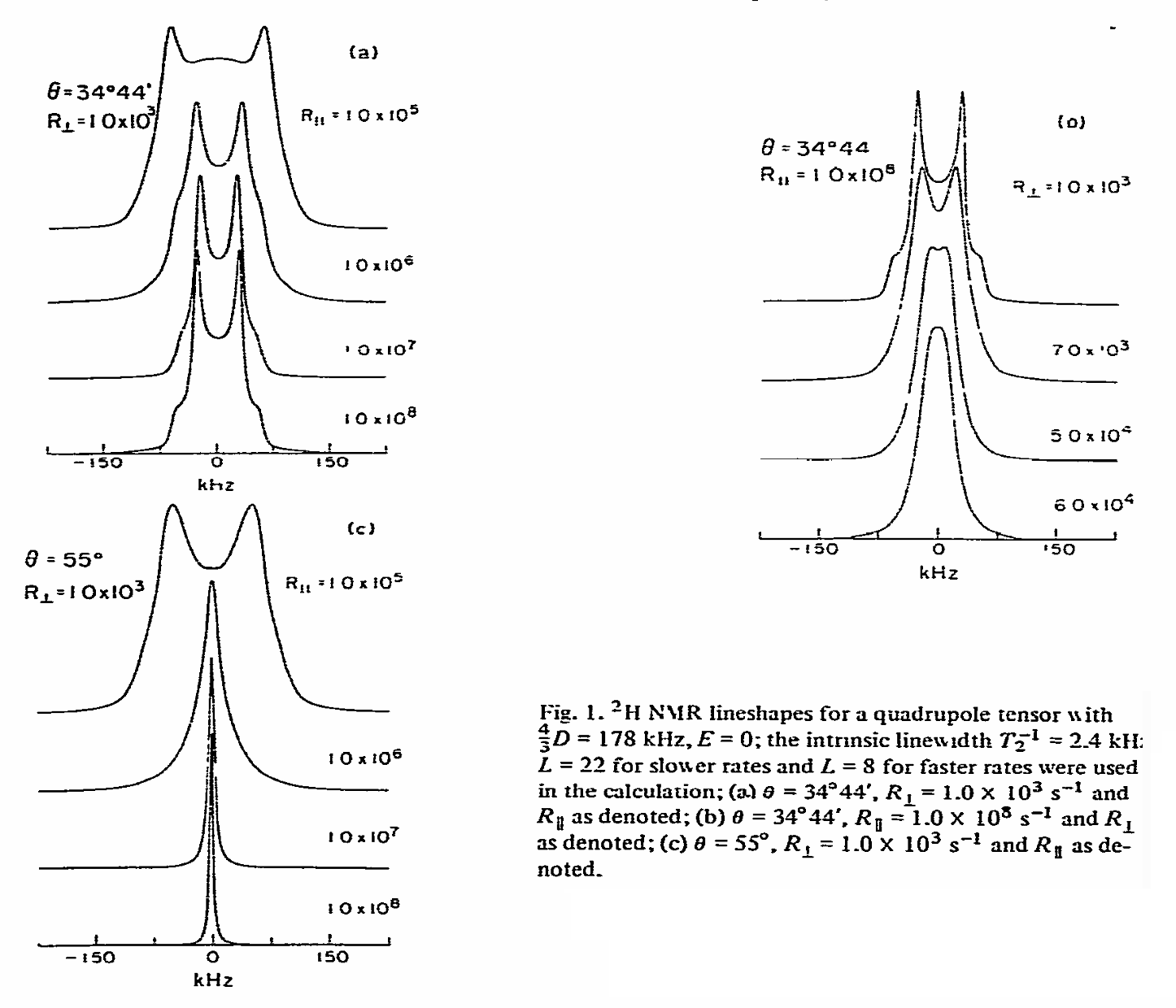
|
|
ABSTRACT: A general method to analyze NMR show motional lineshapes is extended to I = 1 nuclei and illustrated on 2H NMR lineshapes of a clathrate hydrate of tetrahydrofuran (THF-d8). It is shown that "ring puckering" could be the dominant mode of motion for the enclathrated THF-d8 molecule, whereas several other models are inconsistent with experiment.
|
|
|
Chemically-Induced Dynamic Spin Polarization in Two Dimensional Systems: Theoretical Predictions
G.P. Zientara and J.H. Freed.
J. Chem. Phys. 71, 3861-3879 (1979).
<doi: 10.1063/1.438796>
Publication #77
|
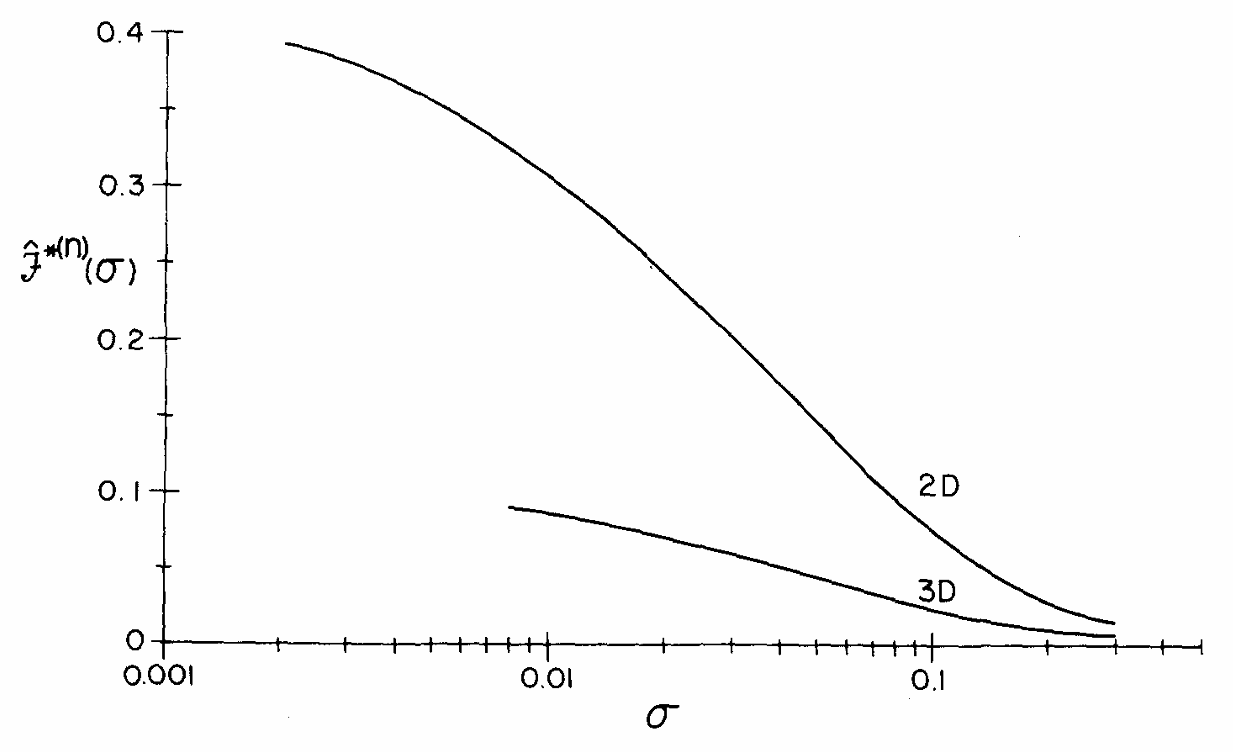
|
|
ABSTRACT: Theoretical predictions for chemically induced dynamic spin polarization [CIDN(E)P] and Heisenberg spin exchange in two dimensional fluid systems are developed. An idealized model, which yields simple limiting results is first discussed in order to illustrate the importance of the geometrical aspects of this problem upon the CIDN(E)P observables. Pedersen–Freed theory, which employs numerical solutions of the stochastic-Liouville equation is then applied. This approach is appropriately modified in its use and analysis in order to fully exhibit the details of two dimensional kinetic and polarization processes. Specifically, the Laplace transformed results are calculated and related to the finite time results (rather than the t→∞ asymptotes) due to logarithmic divergences in time which characterize two dimensional processes. Approximate empirical formulas are developed to describe our exact numerical results and they illustrate the profound effects of a change of dimensionality on the CIDN(E)P observables. These results are related to experimental observables by considering the role of processes which limit the time scale for the polarization (e.g., radical scavenging and T1) and by a consideration of the role of two dimensional bimolecular encounter theory on random initial encounters. Other aspects of two dimensional effects (e.g., time-dependent diffusion coefficients and concentration effects) are briefly noted.
|
|
|
Electron Spin Resonance Study of Anisotropic Rotational Reorientation of Spin Labels Attached to the Side Chains of Soluble Poly(Methacrylamide)-Type Copolymers
J. Pilař, J. Labský, J. Kálal, and J.H. Freed.
J. Phys. Chem. 83, 1907-1914 (1979).
<doi: 10.1021/j100477a024>
Publication #76
|
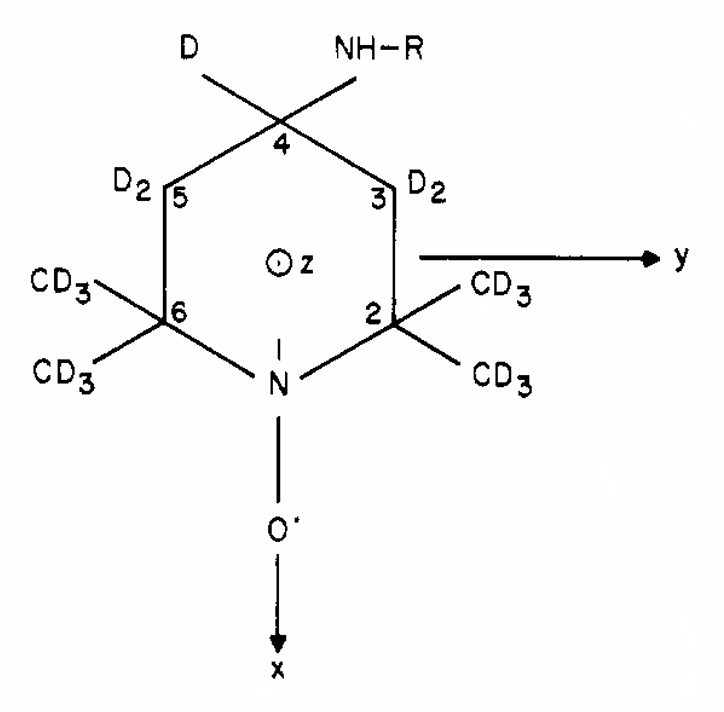
|
|
ABSTRACT: Soluble copolymers of N-(2-hydroxypropyl)methacrylamide (HPMA) with N-methacryloylated ω-amino acids, which form side chains with 4-aminoperdeuterio-2,2,6,6-tetramethylpiperidinyl-l-oxy (PD-Temp-NH2) attached to their end, were investigated. The EPR spectra of methanolic solution of these copolymers recorded at 313-173 K were analyzed by using a model of anisotropic but axially symmetric rotational reorientation of the spin label. An analysis of these spectra in the motional narrowing region showed that the symmetry axis z′ of the rotational-diffusion tensor describing this rotational reorientation was an axis close to that of the N-O bond, and that, depending on the type of side chain, the rotational reorientation about this axis is four to six times faster than about the remaining two axes. The correlation time, τR, characterizing the rate of rotational reorientation of the spin label decreases monotonically with increasing side chain length. Linear log τR vs. 1∕T dependences were obtained and were characterized by a low activation energy, E = 15 ± 2 kJ∕mol, that was the same within the limits of experimental error for all the copolymers. Slow motional spectra were given a preliminary interpretation with a model of very anisotropic rotational reorientation.
|
|
|
The Variational Method and the Stochastic-Liouville Equation. I. A Finite Element Solution to the CIDN(E)P Problem
G.P. Zientara and J.H. Freed.
J. Chem. Phys. 70, 2587-2598 (1979).
<doi: 10.1063/1.437844>
Publication #75
|
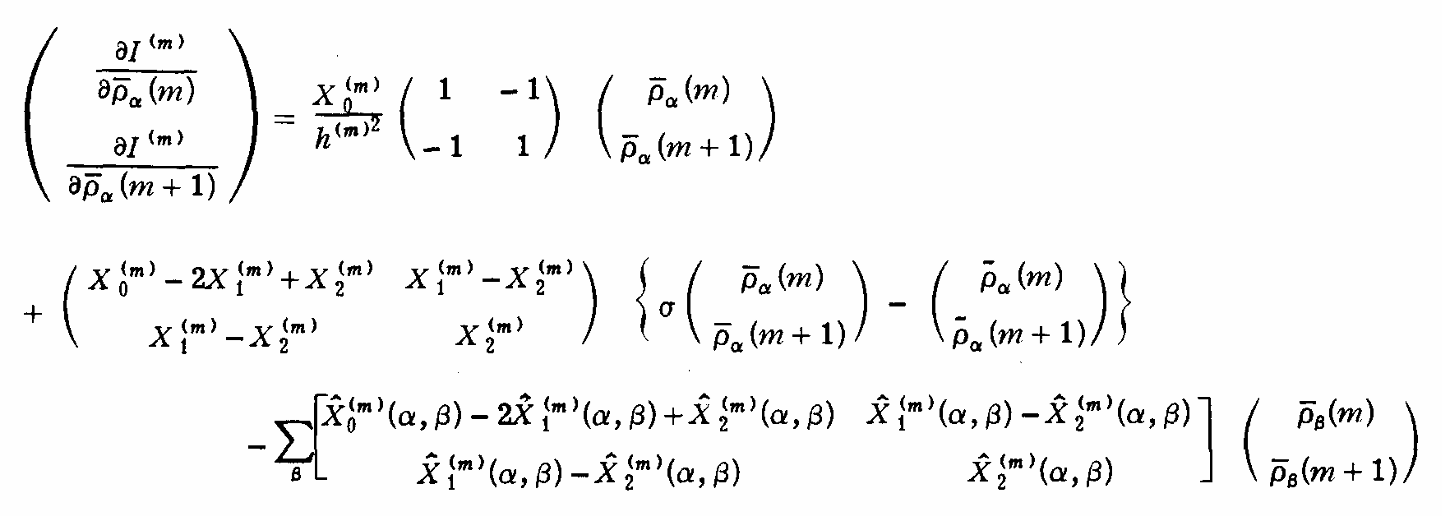
|
|
ABSTRACT: A variational formulation is developed for the stochastic–Liouville equation (SLE). It is shown how this formulation may be used as a general basis for the study of numerical and approximate methods of solution of the SLE. The finite element method is developed for the approximate solution of the spin–density matrix elements using the variational formulation. The method is illustrated by employing it to obtain a compact computer-oriented solution to the (high-field) chemically-induced spin polarization problem. This solution is both more efficient as well as more accurate than the previous treatment by Pedersen and Freed using finite difference methods. Various features of finite element and finite difference methods are compared from the viewpoint of this solution. The great flexibility of finite element methods for solution of the SLE is discussed.
|
|
|
|
|
Theory of Chemically Induced Dynamic Spin Polarization. IV. Low-Field Effects
G.P. Zientara and J.H. Freed.
J. Chem. Phys. 70, 1359-1370 (1979).
<doi: 10.1063/1.437575>
Publication #73
|
|
|
ABSTRACT: The Pedersen–Freed theory of chemically-induced dynamic spin polarization is extended to low field cases where the T±→S transitions must be included. The solutions of the stochastic-Liouville equation are again obtained by numerical finite-difference (FD) methods, but order of magnitude reductions in the size of the matrices to be inverted are realized by introduction of a simple modification of the FD expressions. The simplest case of a single nuclear spin of I=1∕2 on one radical is treated in detail. The results show unequivocally the significant dependence of both CIDNP and CIDEP signals on both the magnitude and the range of the exchange interaction. Also, significant polarizations which should be observable are predicted for both types of experiment under appropriate low field conditions. All the computed results are consistent with a simple qualitative model involving two primary regions in r space: in the outer region the hyperfine terms induce T±→S transitions and J(r)=0, while in the inner region J(r) modulates the effectiveness of these transitions. Consideration is also given to magnetic-field effects on chemical reactivity as well as T±→S contributions in high field but viscous CIDEP.
|
|
|
Slow-Motional NMR Line Shapes for Very Anisotropic Rotational Diffusion. Phosphorus-31 NMR of Phospholipids
R.F. Campbell, E. Meirovitch, and J.H. Freed.
J. Phys. Chem. 83, 525-533 (1979).
<doi: 10.1021/j100467a020>
Publication #72
|
|
|
ABSTRACT: The model of Mason, Polnaszek, and Freed for ESR-slow-tumbling spectra is extended to the case of NMR line shapes for arbitrary tilt of the internal axis of relatively rapid rotation with respect to the principal axes of the chemical shielding tensor for a decoupled I = 1∕2 nucleus. The theory is applied to an analysis of 31P NMR spectra from partially hydrated dipalmitoylphosphatidylcholine (DPPC) molecules over a range of temperature. Generally, very good agreement with experiment is obtained enabling the determination of the rate of the "average" or "effective" internal rotation and the orientation of this "average" axis in the molecule. The basis on which the simple model reflects the spectral effects of the combined internal and overall motions is discussed in detail in an appendix, and the extension to more complex models is outlined.
|
|
|
Theory of ESR Saturation Recovery in Liquids and Related Media
J.H. Freed.
In Time Domain Electron Spin Resonance, L. Kevan and R.N. Schwartz Eds.,
Wiley, 1979; Chapter 2, pp. 31-66.
Publication #71
|
|
|
INTRODUCTION: This chapter is based on the original paper published in 1974, reference 1. It was motivated by the then growing interest in pulsed electron spin resonance (ESR) experiments on free radicals in liquids and related media, in particular saturation recovery-type experiments. Since then the interest has greatly expanded, as is evidenced by this book. The basis of our theory of time-resolved ESR experiments is a natural outgrowth of our theory of steady-state saturation and multiple resonance behavior. This theory has now been summarized in a chapter in another book, and frequent reference is made to it. In fact, it was most interesting to find that the steady-state saturation theory, which has been developed in great detail, could readily be extended, with all its sophistication, to the case of time-dependent, or time-resolved, spectroscopy.
We emphasize saturation recovery in this chapter, but we also include some comments on pulsed electron-electron double resonance (ELDOR), which in principle, may be thought of as a saturation recovery, but with observation at a frequency displaced from the high-power pulse frequency. In later chapters we see how such methods may be extended to free-induction decay and echo-type experiments for the free-radical systems, where numerical techniques are useful. We emphasize analytical techniques in the present chapter. This is possible because one finds that over a wide range of types of systems, the saturation recovery experiment is simply interpreted. We, however, give the general expressions that are amenable to computer methods already developed in connection with steady-state problems.
|
|
|
|
|
Electron Spin Resonance Studies of Anisotropic Ordering, Spin Relaxation, and Slow Tumbling in Liquid Crystalline Solvents. 3. Smectic
W.-J. Lin and J.H. Freed.
J. Phys. Chem. 83, 379-401 (1979).
<doi: 10.1021/j100466a018>
Publication #69
|
|
|
ABSTRACT: In this work the nonaxial ordering and spin relaxation of PD-Tempone spin probe in several liquid crystalline solvents exhibiting smectic A and B phases were studied utilizing methods previously employed by Polnaszek and Freed for the study of nematic liquid crystals. The results reported here for the isotropic and nematic phases are generally in accord with those obtained previously. An analysis of isotropic hyperfine shifts, changes in the ordering tensor, and anomalous relaxation behavior in the smectic phases suggest a model in which the PD-tempone probes are partially expelled from the dipolar region of the liquid crystalline molecules toward the more flexible hydrocarbon end chains as a result of the packing of the smetic layers, and concomitantly the probes increasingly experience a slowly relaxing local structure (SRLS) in a cavity-like location. Differences in observations from different types of smectic liquid crystals are interpreted in terms of their differing structures based on X-ray studies. It is shown that the angular dependent line widths in the smectic phases are significantly affected by the size and shape of the sample. These inhomogeneous broadening effects are discussed in detail in terms of static distortions of the smectic layering induced by wall effects and magnetic-field induced torques, and are in reasonable agreement with predictions of a simple model. The residual homogeneous widths are discussed in terms of combined models of anisotropic rotation and anisotropic viscosity as well as associated SRLS models. For the former case, the problem of defining the rotational diffusion tensor, which must be time dependent in any axis system, is discussed in some detail.
|
|
|
Dynamic Effects of Pair Correlation Functions on Spin Relaxation by Translational Diffusion in Liquids. II. Finite Jumps and Independent T1 Processes
J.H. Freed.
J. Chem. Phys. 68, 4034-4037 (1978).
<doi: 10.1063/1.436302>
Publication #68
|
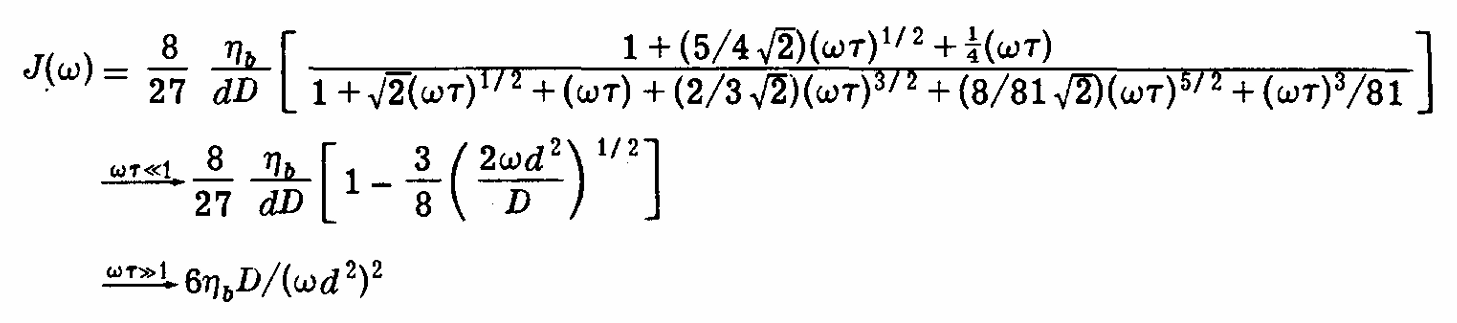
|
|
ABSTRACT: Hwang and Freed have previously given solutions for the relative diffusion of molecules that include the proper boundary condition (i.e., an excluded volume due to a distance of minimum approach) which has usually been neglected in spin relaxation theories. In this work their results are extended to include effects of (1) one type of spin that is rapidly relaxing, (2) diffusion by jumps of finite size, and (3) frequency-dependent diffusion coefficients in the theory of spin relaxation by intermolecular dipolar interactions. These results are mathematically simpler and sounder than those commonly employed. In particular, it is shown that for case (2) measurements of J (O), the zero-frequency spectral density cannot solely be used to determine the jump size, in constrast to the Torrey theory, which did not consider the boundary-value problem.
|
|
|
Electron Spin Relaxation in Isotropic and Ordered Fluids
J.H. Freed.
Proc. XXth Intl. Spect. Colloq., Prague, II, pp. 223-236 (1977).
Publication #67
|
|
|
ABSTRACT: Recent experimental and theoretical studies of spin probes in isotropic and liquid crystalline media are discussed. It is shown how ESR studies of the pressure and temperature dependence of the nematic ordering of 1he probe can be used to test equations of state of this phase and to obtain information on the nature of the orienting potential of the probe. The Freed, Bruno, Polnaszek theory appropriately generalized, is utilized to interpret ESR line shapes and relaxation in the motional narrowing and slow tumbling regions. The nature of reorientational motion under orienting potentials is discussed in terms of anisotropic diffusion and viscosity. It is shown that the effects of the slowly relaxing local structure (SRLS) about the spin probe must be included to explain observed results in boih isotropic and nematic solvents. Such mechanisms can affect the diffusion coefficient, rendering it frequency dependent and/or the mean orienting potential rendering it time-dependent. The properties of the isotropic-nematic phase transitions as probed by ESR is discussed. A general theory combining effects of localized motions and of co-operativity on the spin relaxation is outlined. Recent studies of Heisenberg spin-exchange and relaxation by intermolecular dipolar interactions modulated by translational diffusion are also discussed.
|
|
|
Numerical Methods and Model Dependence in Chemically-Induced Dynamic Spin Polarization
J.H. Freed.
In Chemically Induced Magnetic Polarization, L.T. Muus et al., Eds.
D. Reidel Publishing Company: Dordrecht-Holland, 1977; Chapter 19.
Publication #66
|
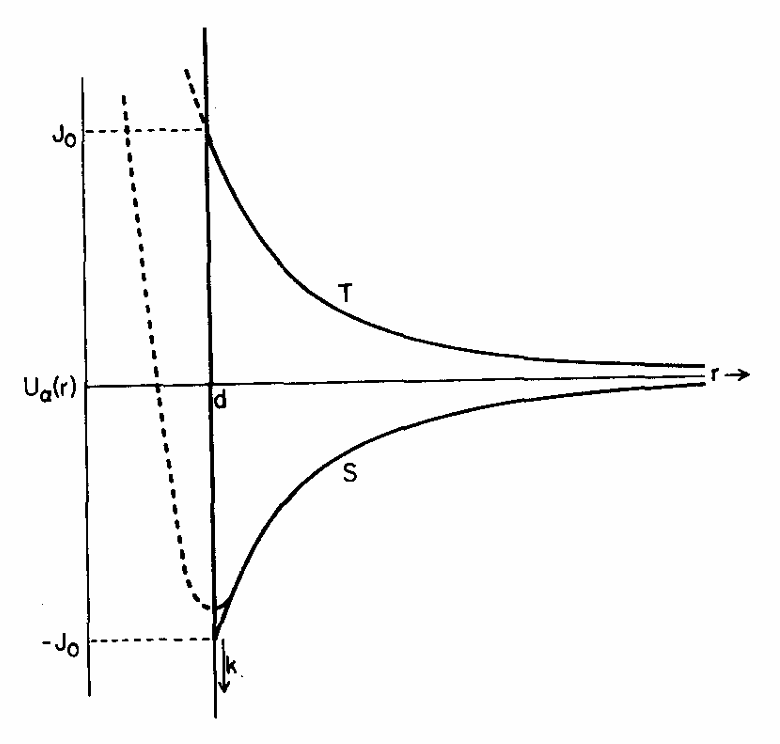
|
|
ABSTRACT: In this chapter we desciibe some of the more advanced theoretical methods, which may be usefully employed in the study of chemically-induced spin polarization and related phenomena. The emphasis will be on the finite difference methods developed by Pedersen and Freed.
|
|
|
Gyromagnetic Ratio
J.H. Freed.
In Encyclopedia of Physics: G.L. Trigg and R. Lerner, Eds.
Addison-Wesley: New York, 1980; pp. 376-377.
|
|
|
Gyromagnetic Ratio
J.H. Freed.
In Encyclopedia of Physics: G.L. Trigg and R. Lerner, Eds.
Addison-Wesley: New York, 1980; pp. 376-377.
Publication #65
|
|
|
INTRODUCTION: The gyromagnetic ratio (or magnetogyric) ratio γ is defined as the ratio of the magnetic moment μ. to the angular momentum J for any system. Specifically, one introduces for electrons with electron spin angular momentum J = ℏS (where ℏ = h∕2π and h is Planck's constant) the electron gyromagnetic ratio γe by μe = −γeℏS, where the negative sign represents the fact that the spin and moment are oppositely directed. For a nucleus, which is a composite particle with total spin I, one introduces the nuclear gyromagnetic ratio γl by μl = γlℏI. Both μl and I (or μe and S) must be regarded as quantum-mechanical operators. In the case of nuclear moments, the defining equation for γl must be considered as implying that the expectation values of μl and I are taken for the given state (usually the ground state) of the nucleus. It then follows from symmetry considerations expressed in the Wigner-Eckart theorem that μl and I may be taken as collinear, with γlℏ the proportionality constant.
|
|
|
Stochastic-Molecular Theory of Spin-Relaxation for Liquid Crystals
J.H. Freed.
J. Chem. Phys. 66, 4183-4199 (1977).
<doi: 10.1063/1.434495>
Publication #64
|
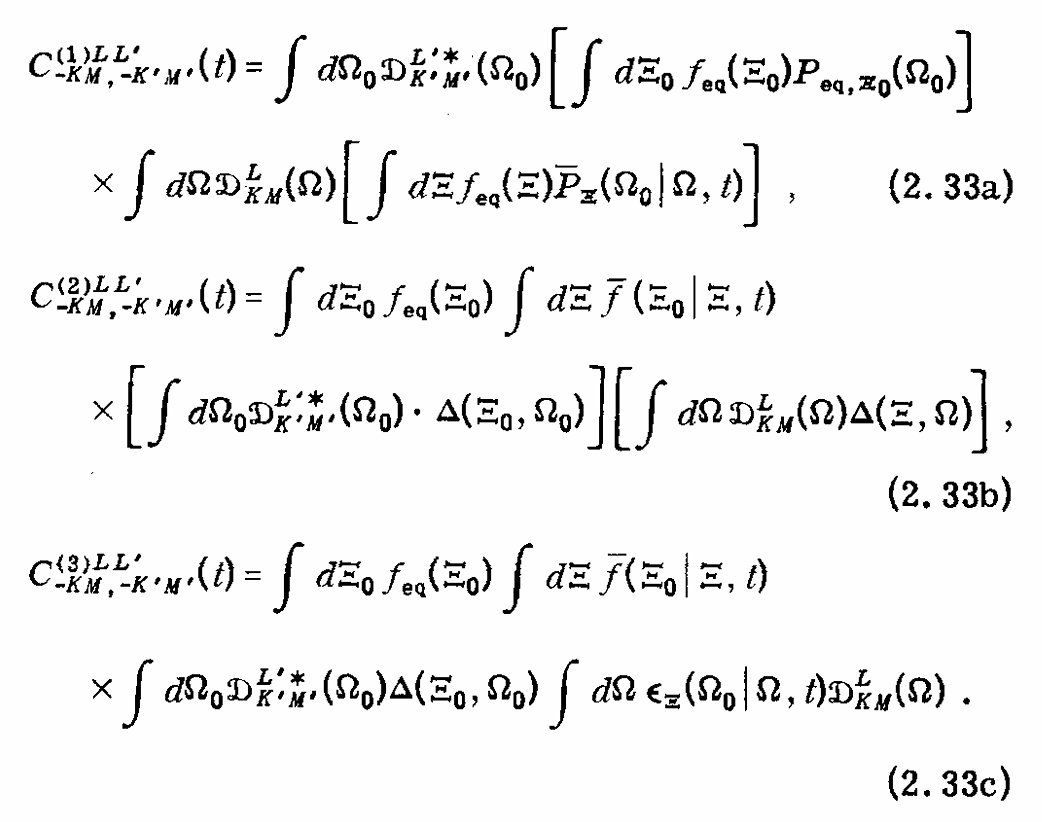
|
|
ABSTRACT: The theory of spin relaxation for liquid crystals is examined with the objective of properly analyzing the statistical interdependence of the faster rotational reorientation of the individual spin-bearing molecules and the (slower) director or order-parameter fluctuations. The analysis is presented in terms of a composite Markov process including both types of motions. It is shown that one recovers a sum of spectral-density terms which, in lowest order in fluctuations, correspond to (1) reorientation of the molecule relative to the equilibrium potential of mean torque, (2) effects of director fluctuations, and (3) a negative cross-term between these two processes which bears a simple relation to (2). Detailed results are given for the particular models of director fluctuations in the nematic phase, quasicritical order fluctuations on either side of the isotropic–nematic phase transition, and slow fluctuations in the local structure. Effects of localized cooperative modes of molecular reorientation are also included. Explicit expressions for NMR and ESR relaxation and line shapes are given. The results obtained here clearly demonstrate some weaknesses in previous treatments which were presumed to be based on an assumption of the statistical independence of the different motional processes. Discussion is also given on how to formulate director fluctuations as a multidimensional Markov process, and on the applicability of motional narrowing theory in these cases where director fluctuations have very slowly relaxing components.
|
|
|
Electron Spin Resonance Studies of Anisotropic Ordering, Spin Relaxation, and Slow Tumbling in Liquid Crystalline Solvents. 2
K.V.S. Rao, C.F. Polnaszek, and J.H. Freed.
J. Phys. Chem. 81, 449-456 (1977).
<doi: 10.1021/j100520a016>
Publication #63
|
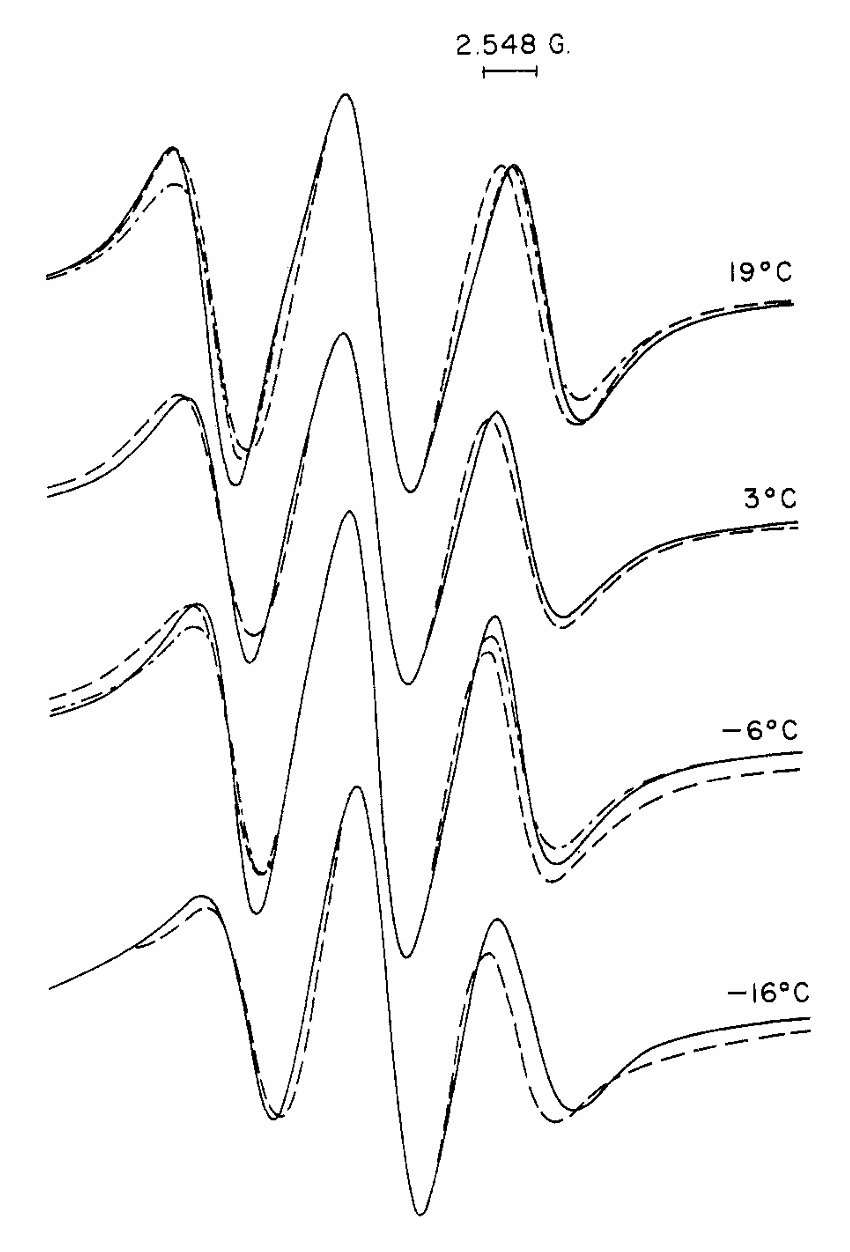
|
|
ABSTRACT: A study is reported of the ESR line shapes in the slow-tumbling region for a cholestane (CSL) spin probe in nematic phase V solution. The line shapes are analyzed in terms of the Polnaszek, Bruno, Freed theory for spectra from slowly tumbling probes in ordered fluids. Rather good agreement with experiment is obtained from the slower motional spectra (τR > 5 × 10−9 s where τR is the rotational correlation time) utilizing a single term ordering potential. For these slow-motional spectra, effects of proton inhomogeneous broadening are relatively small. It is shown in this study on CSL that for (τR > 10−9 s a motional-narrowing theory will lead to erroneous predictions for τR and ordering that become more serious as τR increases. The previous application of motional-narrowing theory by other workers to this probe is discussed in this light. The effects of various other factors upon the spectral simulations are discussed. In particular, it is shown that the spectra for this highly ordered probe should not be very sensitive to various aspects of model dependence of the reorientational motion as discussed in part I nor to effects from director fluctuations. Also the slower tumbling spectra are rather insensitive to anisotropy in τR (but the anisotropy may be determined from the faster motional spectra). Nevertheless, contributions from these effects could modify somewhat the values of τR and ordering that are obtained. The slow tumbling spectra are also insensitive to the angle of tilt of the nitroxide magnetic-tensor principal axis system in the x−y plane with respect to the molecular orientational axes. Our results show that the tilt angle of the magnetic z axis should not be very different from 0°.
|
|
|
ESR Studies of Spin Probes in Anisotropic Media
J.H. Freed.
In Magnetic Resonance in Colloid and Interface Science, H.A. Resing and C.G. Wade, Eds.
ACS Symposium Series 34, pp. 1-15 (1976).
Publication #62
|
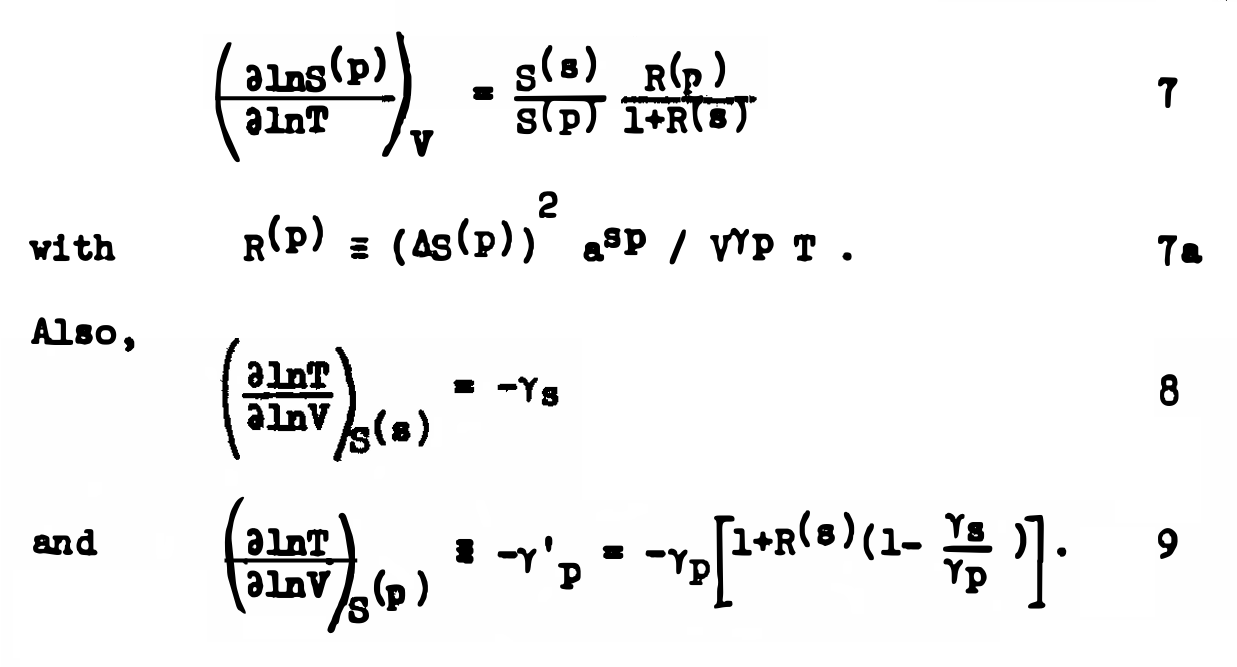
|
|
ABSTRACT: We wish to summarize some of our recent studies of spin probes in liquid crystalline media. We also wish to indicate some theoretical aspects of spin-dependent phenomena on surfaces and interfaces.
|
|
|
Symmetry of Orientational Order Fluctuations about the Nematic-Isotropic Phase Transition: An ESR Study
K.V.S. Rao, J.S. Hwang, and J.H. Freed.
Phys. Rev. Lett. 37, 515-518 (1976).
<doi: 10.1103/PhysRevLett.37.515>
Publication #61
|
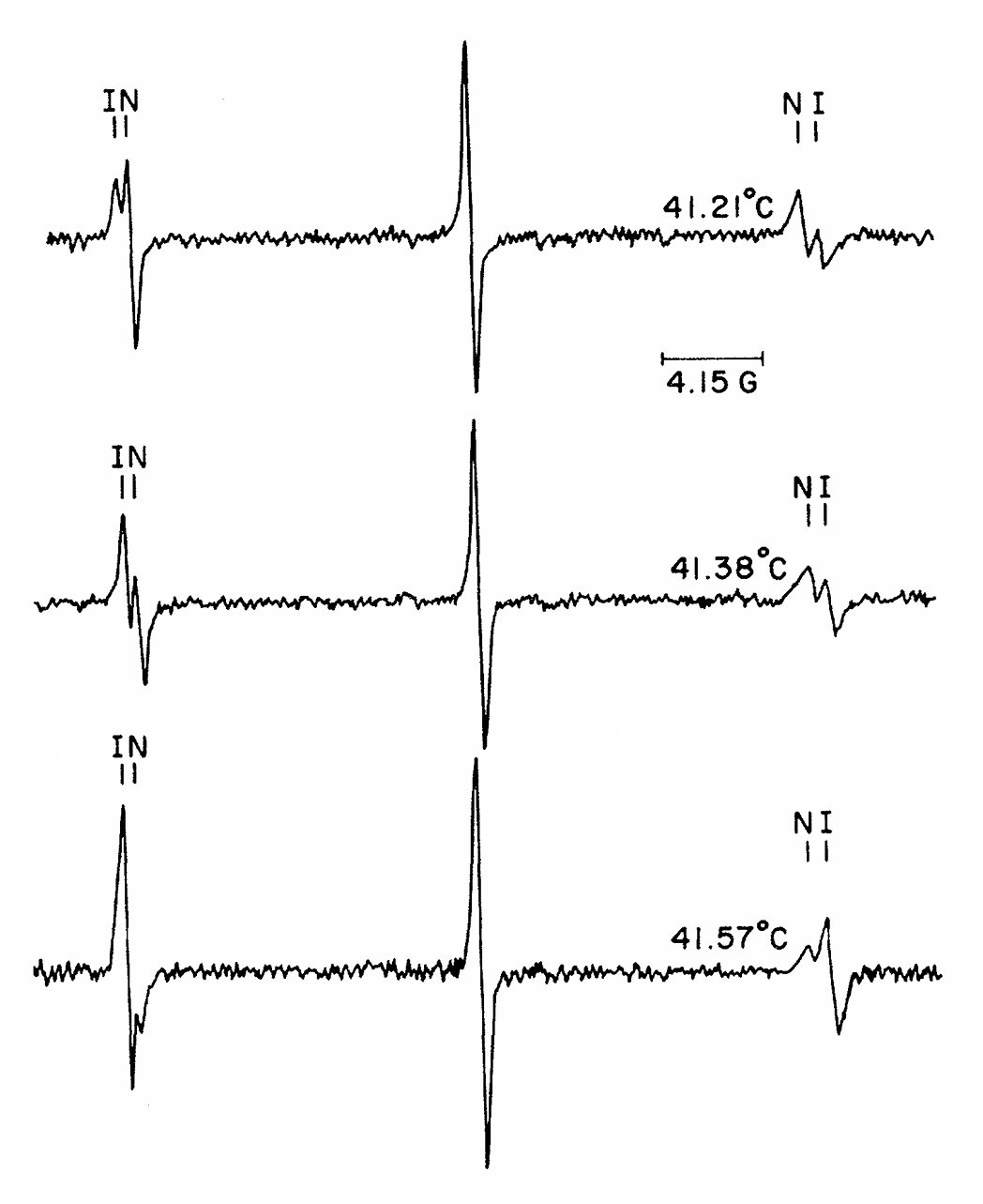
|
|
ABSTRACT: The ESR relaxation of a weakly aligned spin probe dissolved in N-[p-methoxybenzylidine]-p-butylaniline has been studied near Tc, the isotropic-nematic transition. Spin relaxation due to critical orientational fluctuations is observed on either side of Tc and is characterized by a symmetry about Tc that is rather well explained by simple Landaude Gennes mean-field theory for the weak first-order transition.
|
|
|
An Electron Spin Resonance Study of the Pressure Dependence of Ordering and Spin Relaxation in a Liquid Crystalline Solvent
J.S. Hwang, K.V.S. Rao, and J.H. Freed.
J. Phys. Chem. 80, 1490-1501 (1976).
<doi: 10.1021/j100554a019>
Publication #60
|
|
|
ABSTRACT: An ESR study of ordering and relaxation as a function of pressure is reported for the perdeuterated Tempone nitroxide radical probe in phase V solvent. The study of ordering focuses on the nature of the intermolecular interactions between the probe and solvent (and between solvent molecules), which lead to the weak ordering of the probe. The results are suggestive of longer range interactions than were previously found by McColl in an NMR study on a pure solvent. The spin relaxation results have been analyzed in terms of activated-state theory to obtain values of ΔHa and ΔVa as a function of T and P for the rotational reorientation. Anomalous features of the spin relaxation previously reported by Polnaszek and Freed in a temperature-dependent study of this system are reported here as a function of pressure. These anomalies are most pronounced for rotational correlation times (τR) large enough that ESR line shapes are no longer motionally averaged (i.e., they are characteristic of the slow motional region). In general, the anomalous behavior depends mainly on (τR) and appears to be nearly independent of the particular combination of T and P. A mechanism in which the local structure around the probe relaxes more slowly than the probe, as proposed by Polnaszek and Freed, is taken to be the most likely explanation. Rapid motional effects in the rigid solvent are also reported. Details of the high-pressure accessory are given.
|
|
|
Dynamic Effects of Pair Correlation Functions on Spin Relaxation by Translational Diffusion in Liquids
L.-P. Hwang and J.H. Freed.
J. Chem. Phys. 63, 4017-4025 (1975).
<doi: 10.1063/1.431841>
Publication #59
|
|
|
ABSTRACT: It is shown how the equilibrium pair correlation function between spin-bearing molecules in liquids may be incorporated as an effective force in the relative diffusion expressions, and how one may solve for the resulting time correlation functions and spectral densities needed for studies of spin relaxation by translational diffusion. The use of finite difference methods permits the solution no matter how complex the form of the pair correlation function (pcf) utilized. In particular, a Percus–Yevick pcf as well as one corrected from computer dynamics, both for hard spheres, are utilized. Good agreement with the experiments of Harmon and Muller on dipolar relaxation in liquid ethane is obtained from this analysis. Effects of ionic interactions in electrolyte solutions upon dipolar relaxation are also obtained in terms of Debye–Hückel theory for the pcf. Analytic solutions are given which are appropriate for the proper boundary-value problem for the relative diffusion of molecules (i.e., a distance of minimum approach) that has usually been neglected in the spin relaxation theories. Other molecular dynamics aspects of spin relaxation by translational diffusion in liquids are briefly discussed.
|
|
|
Electron Spin Resonance Studies of Anisotropic Ordering, Spin Relaxation, and Slow Tumbling in Liquid Crystalline Solvents
C.F. Polnaszek and J.H. Freed.
J. Phys. Chem. 79, 2283-2306 (1975).
<doi: 10.1021/j100588a015>
Publication #58
|
|
|
ABSTRACT: A detailed study of anisotropic ordering, line shapes, and relaxation is reported for the perdeuterated 2,2,6,6-tetramethyl-4-piperidone N-oxide (PD-Tempone) nitroxide radical in several liquid crystal solvents. The line width results are analyzed in terms of the Polnaszek, Bruno, and Freed (PBF) theory appropriately modified for anisotropic ordering both in the motional narrowing and slow tumbling region. The motional narrowing results are usually consistent with isotropic rotational diffusion, but under a weak (asymmetric) ordering potential, ❬D200❭ -0.1, and activation energies characteristic of the twist viscous properties of the liquid crystal. Anomalous line shape behavior in the incipient slow tumbling region is observed, which is not explained by the extrapolation of the appropriate parameters from the motional narrowing region. This anomaly is discussed in terms of anisotropic viscosity and director fluctuations. The latter is predicted to be of negligible importance for the weakly ordered spin probe, as well as qualitatively of the wrong behavior. Anisotropic viscosity, while apparently able to "explain" the anomaly, leads to physically untenable conclusions. The anomaly is then discussed in terms of slowly fluctuating intermolecular torques, leading to a frequency-dependent diffusion coefficient. While this latter may in part offer an explanation (when one distinguishes between torque components parallel and perpendicular to the director), the implied slowness of the fluctuating torques suggests, from general theory, a new model based upon a local solvent structure around the spin probe which may persist over longer periods than the reorientation time of the spin probe. A simple model calculation of the effects on the ESR relaxation is given. This limiting model is also appropriate for highly structured isotropic liquids. It is shown that such a model could have the same formal spectral effects as anisotropic rotational diffusion, and it would yield non-Debye-like spectral densities of the type that could potentially "explain" the observed incipient slow tumbling anomaly. More general theoretical approaches for the analysis of these effects are briefly discussed.
|
|
|
The Theory of Chemically Induced Dynamic Spin Polarization
J.H. Freed and J.B. Pedersen.
Adv. Magn. Reson. 8, 1-84 (1976).
Publication #57
|
|
|
INTRODUCTION: In this paper we wish to present the theory for the very interesting phenomenon of chemically induced dynamic spin polarization. We shall consider both the NMR case of chemically induced dynamic nuclear (spin) polarization (CIDNP) and the ESR case of chemically induced dynamic electron (spin) polarization (CIDEP). This subject, which involves a combination of magnetic resonance and spin-selective reaction dynamics in liquid solution, now has a large literature assiciated with it including reviews and books. We will not try to summarize this literature here; the reader is referred instead to the other sources especially for the extensive experimental results. We offer a single coherent and unified treatment of the main theory. That is, we approach the problem from a very general formulation, known as the stochastic Liouville equation (SLE), in which both the spin dynamics and the diffusive and reactive dynamics can be treated simultaneously and in great detail. The many earlier simplified theoretical analyses of the radical-pair mechanism (RPM) are in fact found to be based upon simplified submodels, which are all naturally included in precisely their correct relative importance in the general treatment.
|
|
|
Generalized Einstein Relations for Rotational and Translational Diffusion of Molecules Including Spin
L.-P. Hwang and J.H. Freed.
J. Chem. Phys. 63, 118-130 (1975).
<doi: 10.1063/1.431064>
Publication #56
|
|
|
ABSTRACT: In this work the validity of generalized Einstein relations, D(ω) = kT ∕ Ibeta;(ω), where D(ω) and beta;(ω) are frequency-dependent diffusion and damping coefficients, is examined from a general point of view. It is shown that generalized Smoluchowski (S) equations for both translational and rotational diffusion involving the D(ω), which are defined in terms of simple correlation functions of the fluctuating forces and torques, follow from the appropriate generalized Fokker–Planck (FP) expressions when the translational and angular momenta are taken as rapidly relaxing and a coarse graining in time is introduced. It is found useful in this context to distinguish between those forces and torques fluctuating at rates faster than the diffusive-type motions of the B particle versus those fluctuating more slowly. The appropriate FP equation for rotational diffusion is also derived. Generalized FP and S equations are also derived for the semiclassical case where the molecule(s) examined contains spin degrees of freedom. A proper application of the correspondence principle leads to certain terms referred to as "spin-force" or "spin-torque" terms which have the property of tending to restore the spins to their thermal equilibrium value, an important feature usually lacking in semiclassical treatments. Some simple examples of the generalized S equations are given in terms of memory function approximations of the random force and torque correlation functions. The resulting expressions are seen to bear a close formal similarity to typical expressions developed for simple jump models in either orientational or translational space. It is suggested that recent frequency-dependent experiments, including some previously interpreted in terms of simple jump models, may be amenable to analysis in terms of generalized Einstein and S equations.
|
|
|
A Comparison of Generalized Cumulant and Projection Operator Methods in Spin-Relaxation Theory
B. Yoon, J.M. Deutch, and J.H. Freed.
J. Chem. Phys. 62, 4687-4696 (1975).
<doi: 10.1063/1.430417>
Publication #55
|
|
|
ABSTRACT: The general spin-relaxation theories of Albers and Deutch and of Argyres and Kelley based on different projection operator methods, and the theory of Freed based on generalized cumulant expansions are compared. It is shown that the first two yield equivalent expressions for the time evolution of the spin density matrix. They are also found to be equivalent to a cumulant expansion based on total ordering of the cumulant operators (TTOC), which is different from the partial time ordering method (PTOC) used by Freed. The TTOC method is found to be the more convenient for the frequency domain (i.e., for calculating spectra), while the PTOC method is for time domain analyses. Examples of the use of the TTOC method are given. Useful expressions are given for the case where the lattice may be treated in terms of classical Markov processes, but, in general, it is found that for such cases the stochastic Liouville method is the more useful for computations.
|
|
|
An ESR and ENDOR Study of Spin Relaxation of Semiquinones in Liquid Solution
D.S. Leniart, H.D. Connor, and J.H. Freed.
J. Chem. Phys. 63, 165-199 (1975).
<doi: 10.1063/1.431042>
Publication #54
|
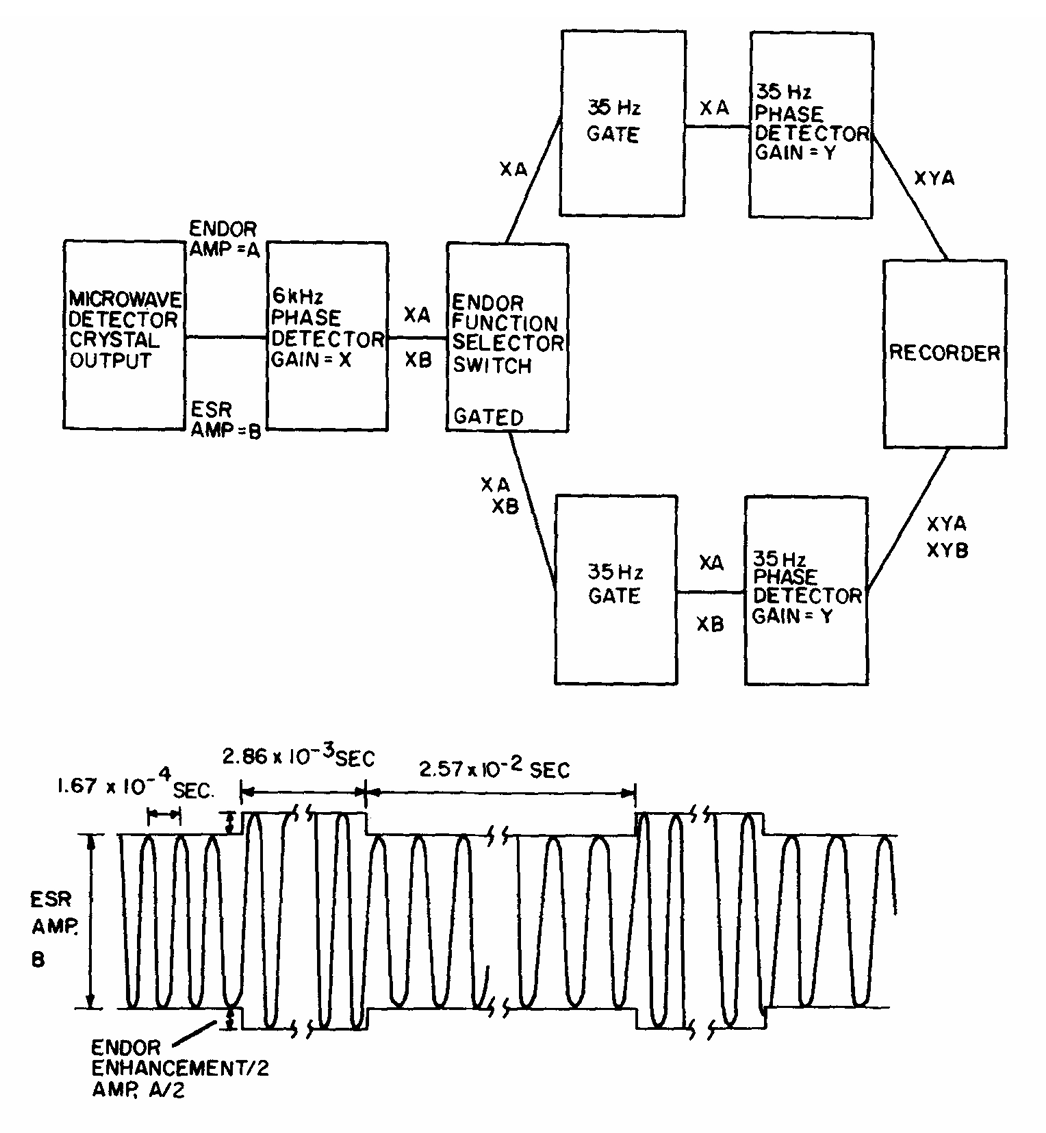
|
|
ABSTRACT: A quantitative test of the ENDOR theory for free radicals in solution has been developed experimentally using the semiquinone (SQ) radical anions of parabenzo- (PBSQ), duro- (DSQ), and 2,5-dimethyl-para-benzo- (2,5-DPMBSQ) dissolved in ethyl alcohol (EtOH) and dimethoxyethane (DME) solvents. It is shown that, in general, an ENDOR signal arising from a molecule containing four or less equivalent nuclei, such as PBSQ, can be analyzed rigorously whereas a molecule containing more than four equivalent nuclei, such as DSQ, which, in principle, could be analyzed rigorously, practically is best analyzed using approximate forms of the ENDOR theory. It is shown that 2,5-DMPBSQ may be analyzed using a combination of both the rigorous and approximate forms of the ENDOR theory. The analysis involves calculation of the ENDOR relaxation parameters, T2n, Ωn, and Ωe,n, from the experimental ENDOR percent enhancement and linewidth studies. A comparison was made by performing the ESR linewidth and saturation studies to obtain values of We, the electron spin–lattice relaxation probability, and Wn, the nuclear spin-relaxation probability and then using this information to predict the observed ENDOR relaxation parameters in terms of the theory. The over-all behavior of the experimental results was found to be fully consistent with the general trends predicted by the theory and expected from the quantitative measurements obtained from the ESR studies. This work describes in detail the instrumentation and experimental methods necessary to measure relaxation parameters of organic free radicals in solution. A comprehensive section describing the effects of modulation amplitude, microwave power, coherence broadening or splitting, and pulse rate is given along with methods for analyzing ENDOR line shapes in their presence. The results and analyses of all ESR and ENDOR experiments are described in detail. Analysis of the rotational correlation time τR, obtained from the ESR studies, shows the radical anions in DME solution are most likely solvated and affected by the counterion and/or supporting electrolyte while PBSQ in EtOH is most likely dissociated. A comparison of the experimental and theoretical magnitudes of We leads to the possibility that for small molecules in liquids a Brownian model of reorientation by infinitesimal jumps need not be satisfactory and that jumps of large angle would be needed to better fit the experimental data. The effect of Heisenberg exchange on ENDOR signals is analyzed and discussed in detail, leading to the conclusion that the intermolecular electron–electron dipolar interaction as well as Heisenberg exchange make contributions to the concentration dependent portion of the linewidth. However, the latter contributes much more to the ESR linewidths as compared to the ENDOR widths for PBSQ in EtOH. The above analysis is modified to include charge effects. Finally it is shown that for methyl group ENDOR cross relaxation from modulation of the isotropic hyperfine splitting via methyl group rotation is not the dominant nuclear relaxation mechanism and all methyl group ENDOR analyses are consistent with a Wn derived from the pseudosecular terms of the anisotropic electron–nuclear dipolar interaction.
|
|
|
Theory of Chemically Induced Dynamic Electron Polarization. III. Initial Triplet Polarizations
J.B. Pedersen and J.H. Freed.
J. Chem. Phys. 62, 1706-1711 (1975).
<doi: 10.1063/1.430695>
Publication #53
|
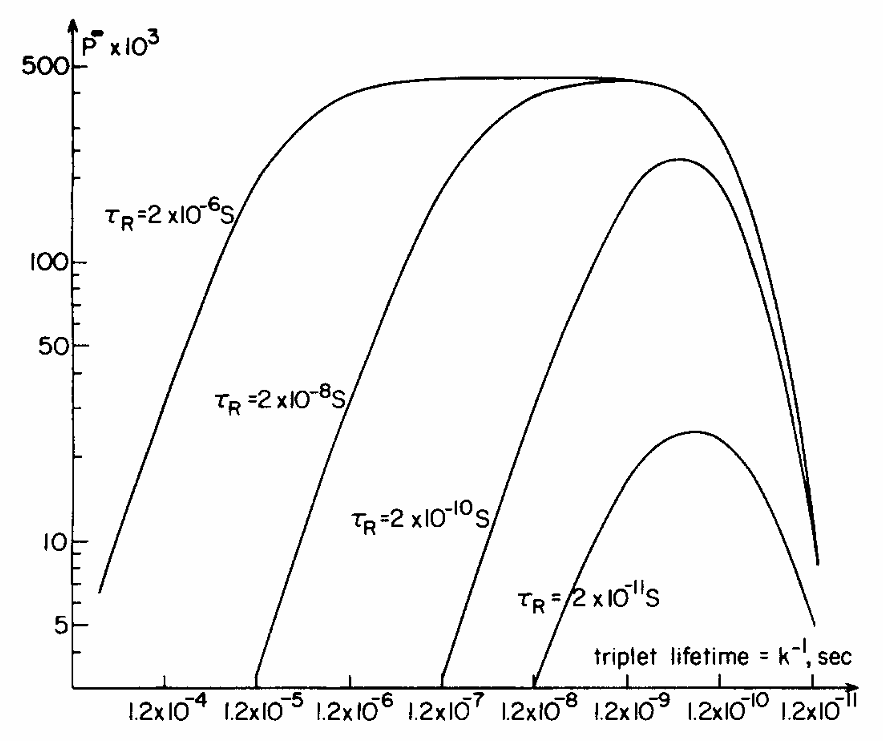
|
|
ABSTRACT: An analysis of dynamical aspects of the CIDEP mechanism proposed by Wong et al. is given.This mechanism is based upon the formation of an excited triplet by intersystem crossing that populates the three triplet levels unequally. The subsequent rotational averaging of the initial population distribution coupled with the orientational effects of the zero field splitting is carefully treated in this work utilizing the stochastic Liouville equation in a manner closely analogous to that recently given for ESR line shapes and relaxation of slow-tumbling triplets. It is shown that the predicted CIDEP polarizations can indeed be very substantial, in agreement with Wong et al., but they will depend in general on the relative magnitudes of not only the zero field (D and E) and Zeeman terms (ω0) but also the relevant reaction rates and the rotational tumbling times (τR). A useful perturbation expression valid for D2 ≲ (1∕2)[ω02 + τR−2] is obtained which shows these details. Typical complete solutions, obtained numerically, are given for cases when this inequality does not hold.
|
|
|
Electron Spin Resonance Studies of Anisotropic Rotational Reorientation and Slow Tumbling in Liquid and Frozen Media. III. Perdeuterated 2,2,6,6-Tetramethyl-4-piperidone N-Oxide and An Analysis of Fluctuating Torques
J.S. Hwang, R.P. Mason, L.-P. Hwang, and J.H. Freed.
J. Phys. Chem. 79, 489-511 (1975).
<doi: 10.1021/j100572a017>
Publication #52
|
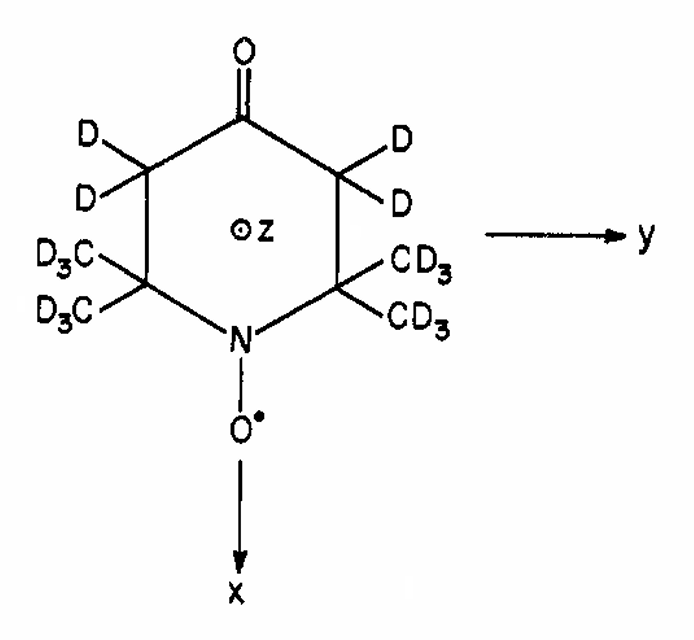
|
|
ABSTRACT: A detailed analysis of line shapes and relaxation is made for the perdeuterated 2,2,6,6-tetramethyl-4-piperidone N-oxide (PD-Tempone) nitroxide radical covering the whole range from fast motional narrowing (τR ˜ 10−12 sec) to the rigid limit (τR ˜ 10−6 sec) in several deuterated solvents. The slow tumbling results show particularly good agreement with the slow tumbling theory of Freed, et al., for a reorientational model of moderate jumps (ca. 50° rms), and essentially isotropic reorientation. It is shown that while such a model may not be readily distinguished, purely from its slow tumbling spectral predictions, from a free diffusion model including inertial effects, the latter model is incompatible with most other considerations. However, a simple analysis explicitly including the fluctuating (or random) torques indicates that more fundamental analysis of the dynamics may explain the slow tumbling results without necessarily involving substantial jumps. The spectral analysis is considerably enhanced by the increased resolution obtained from the use of the perdeuterated spin probe and deuterated solvents. Careful analysis of results at X-band and 35 GHz has shown that the nonsecular spectral densities exhibit significant deviations from a Debye-like spectral density yielding results very similar to those recently reported for the peroxylaminedisulfonate (PADS) radical. Related observations are discussed of apparent non-Debye-like spectral densities in the incipient slow tumbling spectra. These anomalies are also found to be amenable to a unified explanation in terms of the fluctuating torques. The analysis of much of the results in terms of a simple model yields an rms value for the fluctuating torques of ca. √(6)kT and τM &tilde τR where τM is the relaxation time of the torques. The high-temperature electron-spin flip processes are consistent with a Hubbard-type spin-rotational mechanism, but the low-temperature results may be due to spin-rotational relaxation from intramolecular motions. The simple analysis of spin-rotational relaxation in terms of relaxation of the fluctuating torques is found to yield equivalent predictions to conventional treatments when the estimated values of rms torque and τM are used.
|
|
|
A Hydrodynamic Effect on Chemically Induced Dynamic Spin Polarization
J.B. Pedersen and J.H. Freed.
J. Chem. Phys. 62, 1790-1795 (1975).
<doi: 10.1063/1.430704>
Publication #51
|
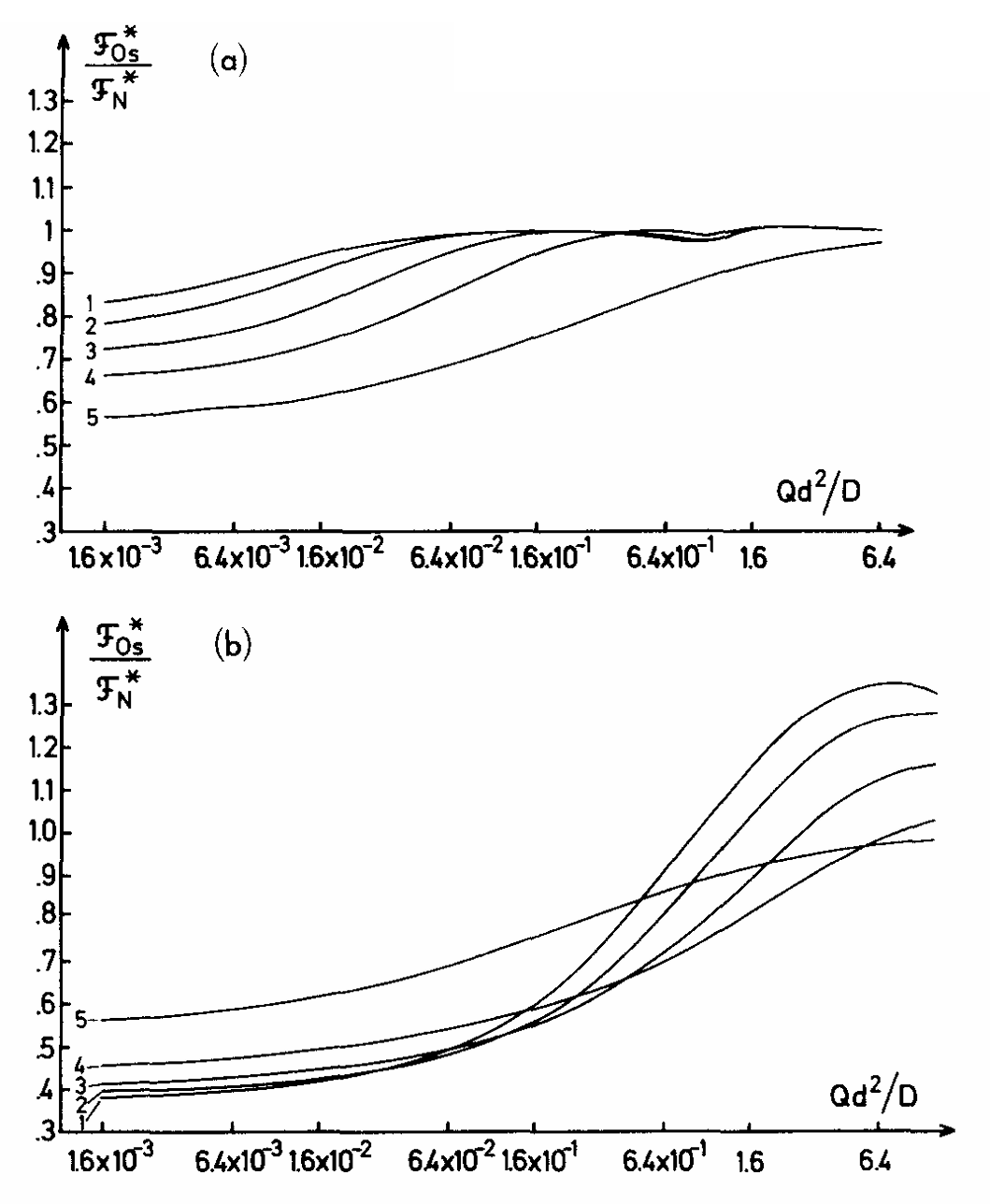
|
|
ABSTRACT: Deutch has recently suggested that the effect of the hydrodynamic interaction represented by Oseen's tensor, on the relative diffusion of radical pairs, should have consequences for CIDNP and CIDEP signals. Detailed solutions to this problem have now been obtained by the finite difference approach of Pedersen and Freed to the stochastic−Liouville equation for the radical−pair mechanism. These solutions show that the hydrodynamic interaction typically leads to polarizations enhanced by factors of the order of 0.9−3 for CIDEP and reduced by factors of (3∕4) −2(1∕2) for the appropriate CIDNP parameter, and these factors are a function of the nature and magnitudes of the various interactions.
|
|
|
Some Theoretical Aspects of Chemically Induced Dynamic Nuclear Polarization
J.B. Pedersen and J.H. Freed.
J. Chem. Phys. 61, 1517-1525 (1974).
<doi: 10.1063/1.1682096>
Publication #50
|
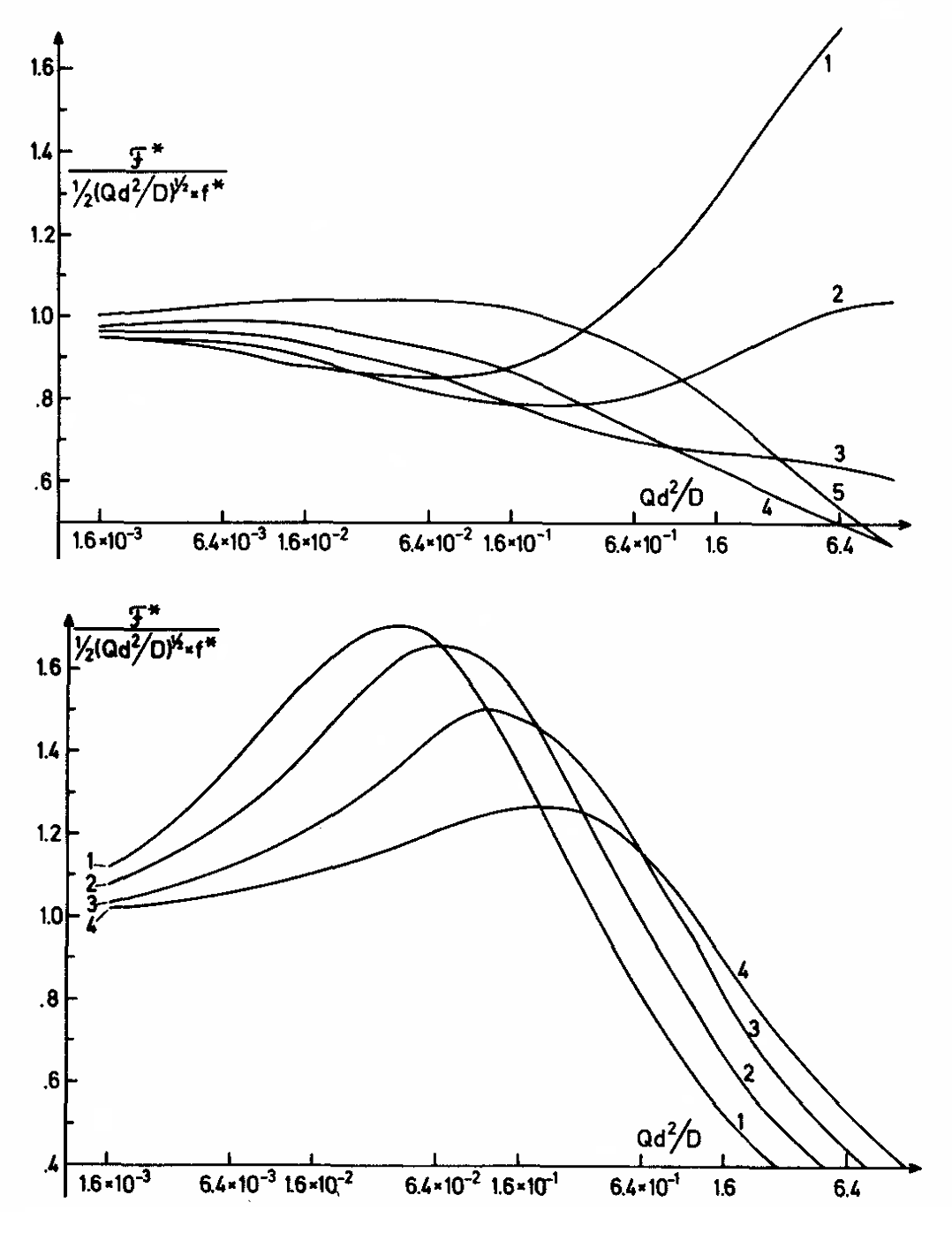
|
|
ABSTRACT: An analysis of aspects of the theory of chemically induced dynamic nuclear polarization (CIDNP) is given in terms of rigorous numerical solutions to the stochastic Liouville equation, in accordance with the methods previously developed for CIDEP. This analysis includes not only a model in which the exchange interaction is of finite extent in the spin Hamiltonian (EFA model) but models in which the exchange interaction explicitly affects the reactive trajectories (EFP model) by their inclusion in a spin-dependent diffusion equation and in which charge interactions between reacting ionic radicals and their surroundings are accounted for in the Debye-Hückel fashion. Several useful and simple relationships are found for the CIDNP phenomenon, and their dependence upon the model is discussed. It is found that the CIDNP polarizations are readily described in terms of two fundamental parameters-Λ, the spin-independent probability of reaction of singlets per collision, which includes all re-encounters, and F∗, which measures the conversion from triplets to singlets for the whole collision. Exact relations for the CIDNP polarizations are given in terms of these two parameters and are found to be nearly independent of model. The parameter Λ is shown to be simply related to k(r), the singlet reaction rate when the radicals are in contact, and to τ1, the effective lifetime of the reacting pair. Simple expressions for τ1 are given for all the models, and these results are compared with those for the usual discussions of diffusion rates on chemical kinetics. It is found that for normal diffusion rates, F∗ obeys the relation (1∕2)√(Qτd), very similar to that first proposed by Adrian, where Q is half the difference in resonant frequencies of the radicals, and τd =d2∕D with D the relative diffusion coefficient and d the distance of closest approach. This relation is not appropriate for viscous systems, and the correct results are given for such cases. The effects of the finite range of the exchange interaction and the longer range Coulombic interactions between radicals upon F∗ for the different models are also obtained. In particular, for EFA the finite range of exchange yields an "excluded volume" effect wherein singlet-triplet mixing (or Q mixing) is ineffective. The model dependent effects upon F∗ are closely related to the recurrence probabilities, and further results are obtained implying a simple expression for the first encounter probabilities of separated radicals under the effects of the different interactions. The polarizations are related to the time-dependent CIDNP intensities that one may observe for a typical scheme of radical production, reaction, and relaxation.
|
|
|
Theory of Slow Tumbling ESR Spectra for Nitroxides
J.H. Freed.
In Spin Labeling Theory and Applications, L.J. Berliner, Ed.
Academic Press: New York, 1976; Chapter 3.
Publication #49
|
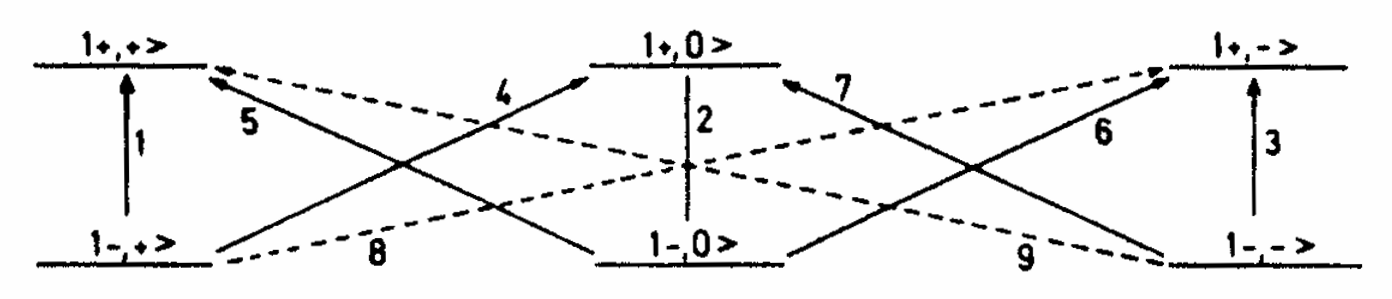
|
|
INTRODUCTION: In this chapter we develop the theory for slow tumbling in ESR spectroscopy, with specific application to nitroxide free radical spectra. The slow tumbling region is that range of rotational reorientation times for which the ESR spectrum can no longer be described as a simple superposition of Lorentzian lines, characteristic of the fast motional or motional narrowing region, for which the theory developed in the previous chapter applies. In this chapter we further take it to be the region for which the ESR spectrum still shows effects of the motion; i.e., the motion is not so slow as to yield a proper rigid-limit spectrum. For nitroxide free radicals, this usually means we are considering the range of rotational correlation times 10−9 sec ≦ τR ≦ 10−6 sec. This is an important range for nitroxide probes in viscous media or for nitroxide spin labels attached to large macromolecules.
In this region, the spectra are affected in a complicated way by both the motions and the magnetic spin interactions. As a result, a theory which can deal rigorously with describing this region must be both powerful and general. It is our objective to set forth this theory, which has been developed in the last few years, and to emphasize its general foundations. The general theory is presented in Section II.A. It is a characteristic of the slow motional region that it requires that we ask more intricate questions about the detailed nature of the molecular motions in order to properly analyze the spectra than is true for studies in the motional narrowing region. Thus in much of the remainder of Section II the dynamics of molecular motions is developed to an extent concomitant with the need to explain actual slow tumbling experiments.
|
|
|
Comments on the Interpretation of Electron Spin Resonance Spectra of Spin Labels Undergoing Very Anisotropic Rotational Reorientation
R.P. Mason, C.F. Polnaszek, and J.H. Freed.
J. Phys. Chem. 78, 1324-1329 (1974).
<doi: 10.1021/j100606a016>
Publication #48
|
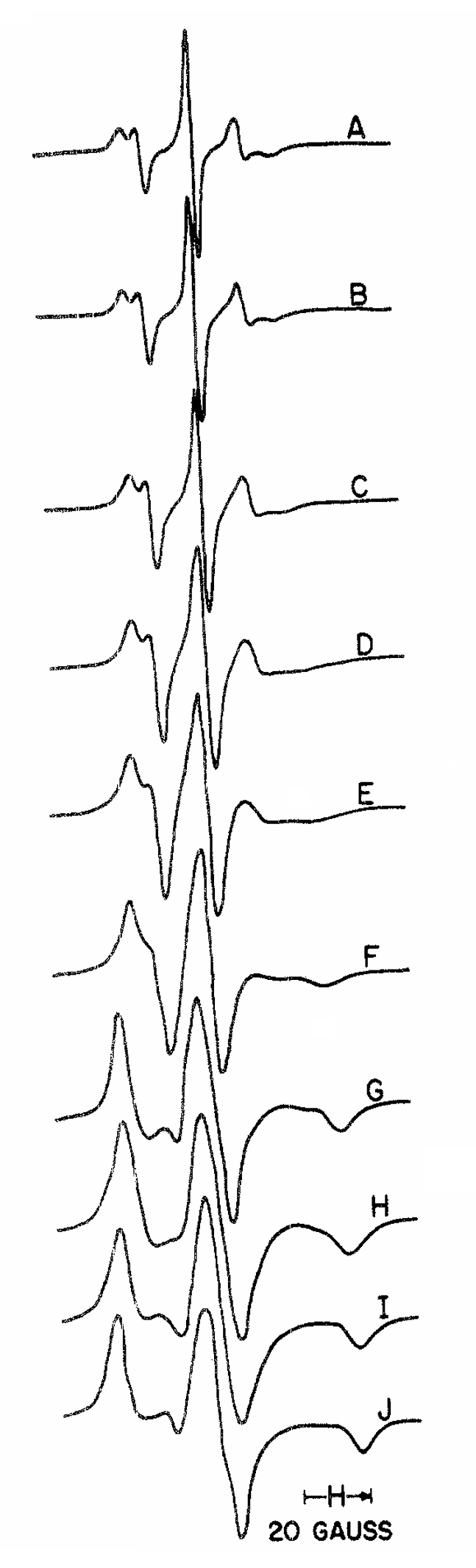
|
|
ABSTRACT: An analysis is given for motional effects on esr spectra of spin labels undergoing very anisotropic rotational relaxation in isotropic liquids. This analysis examines the spectral consequences for cases when the nitroxide is undergoing rapid rotation about a single bond, while the macromolecule to which it is attached is reorienting slowly. A simplified effective diffusion tensor R is employed such that R∥ refers to the bond motion and R⊥ to the effects of overall rotation. This approach is seen to be a useful approximation to developing the motional corrections to the effective time-independent S parameter approach of McConnell, et al. Our analysis including motional effects is found to be in good agreement with recent experiments of Wee and Miller on spin-labeled polymers. We show how motional effects may account for certain inconsistencies of results on polymer and membrane studies originally interpreted in terms of the effective time-independent approach, and how motional effects can influence the interpretation of the S parameter.
|
|
|
Theory of Saturation and Double Resonance in Electron Spin Resonance Spectra. VI. Saturation Recovery
J.H. Freed.
J. Phys. Chem. 78, 1155-1167 (1974).
<doi: 10.1021/j100605a006>
Publication #47
|
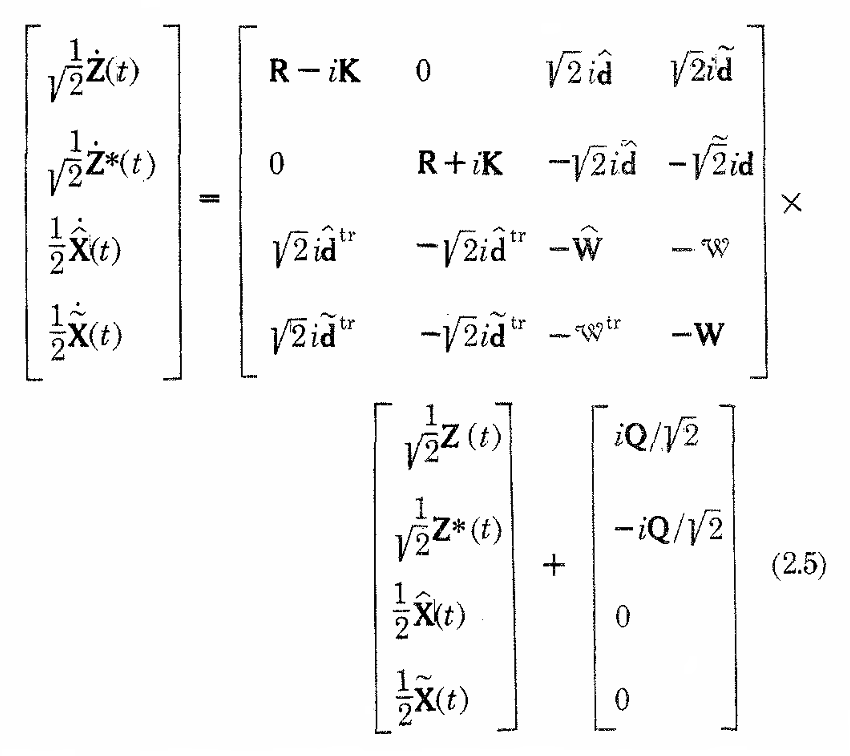
|
|
ABSTRACT: The general theory of Freed for steady-state saturation and double resonance in esr spectra of free radicals is extended to cover time-dependent experiments. The solution is again found to depend on the same matrix representations developed in the earlier work. Particular attention is paid to saturation recovery in the light of recent such experiments. It is shown that while the general solutions yield sums of many exponential decays, the dominant observation may, to a first approximation in many cases, be described in terms of single T1 = ½We, where We is the electron-spin flip rate, in agreement with recent observations. This T1 is characteristically the slowest decay constant, and is either well separated from the much faster decays (due to nuclear spin flip, exchange, and∕or reorientational effects), or may be difficult to distinguish from decay constants of comparable magnitude (due to the same type of effects). Both conventional saturation recovery and eldor-type recoveries are discussed from this point of view. The general approach given is equally adaptable to cases of esr spectra in the motional narrowing region as well as esr spectra characteristic of slow tumbling. Both cases are discussed in detail with several examples given. In particular, in the slow tumbling region for the case of a radical with hyperfine structure (e.g., a nitroxide), it is shown that, in general, both direct reorientational effects as well as nuclear spin flip processes contribute directly to the relaxation modes. The analysis given emphasizes analytic aspects although the general expressions, appropriate for accurate computer simulation, are given.
|
|
|
ESR Lineshapes and Saturation in the Slow Motional Region: ELDOR
G.V. Bruno and J.H. Freed.
Chem. Phys. Lett. 25, 328-332 (1974).
<doi: 10.1016/0009-2614(74)85309-1>
Publication #46
|
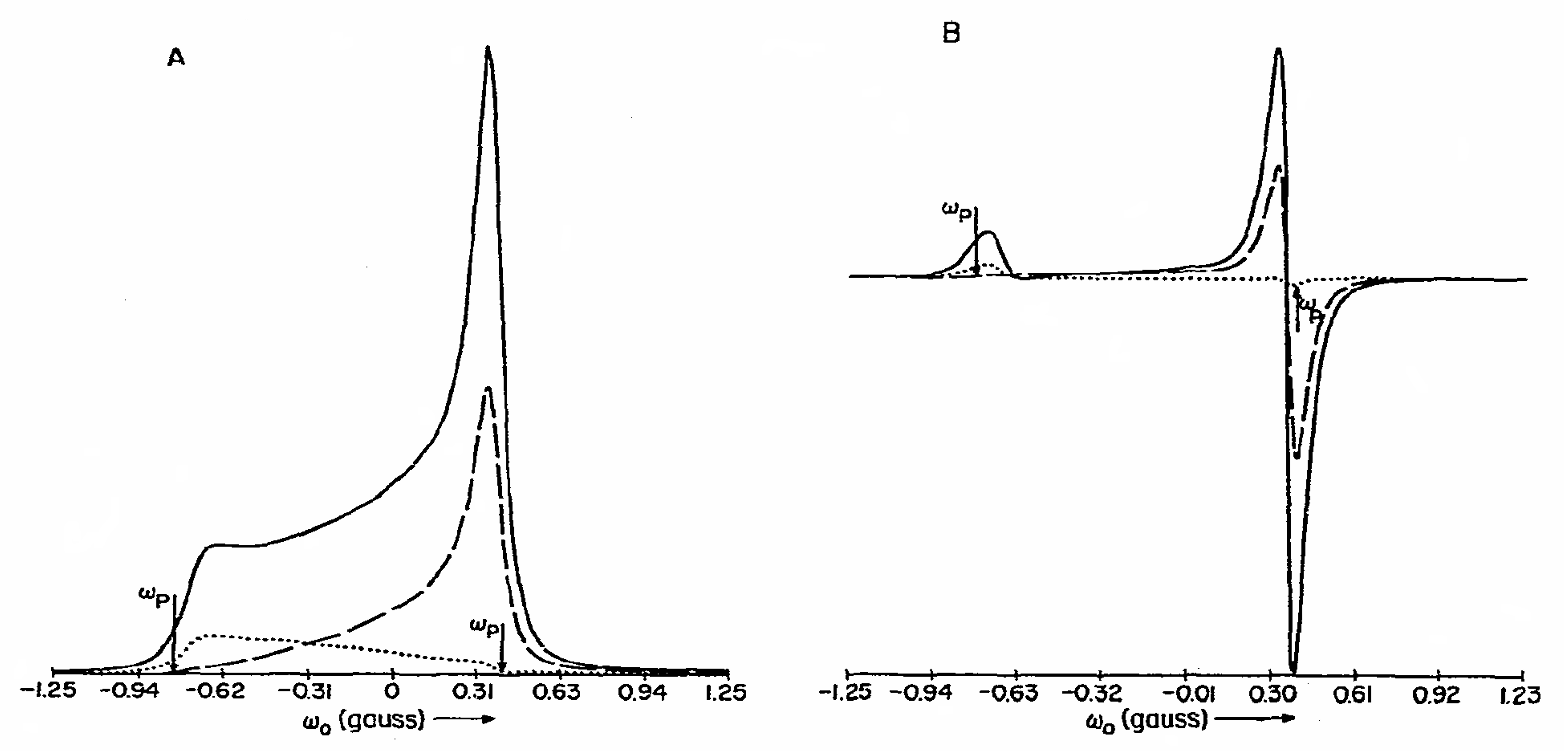
|
|
ABSTRACT: It is shown how the stochastic Liouville approach of Freed, Bruno, and Polnaszek for describing ESR lineshapes in the slow motional region may be applied to the ELDOR experiment. It is possible, in the slow motional region, to pump with a saturating microwave field at one resonant frequency corresponding to a particular orientation and observe effects from the rotational motion in transmitting the saturation to another resonant position. Some effects are illustrated by simulation of ELDOR spectra for a simple ESR line in the slow motional region.
|
|
|
Analysis of Inertial Effects on Electron Spin Resonance Spectra in the Slow Tumbling Region
G.V. Bruno and J.H. Freed.
J. Phys. Chem. 78, 935-940 (1974).
<doi: 10.1021/j100602a015>
Publication #45
|
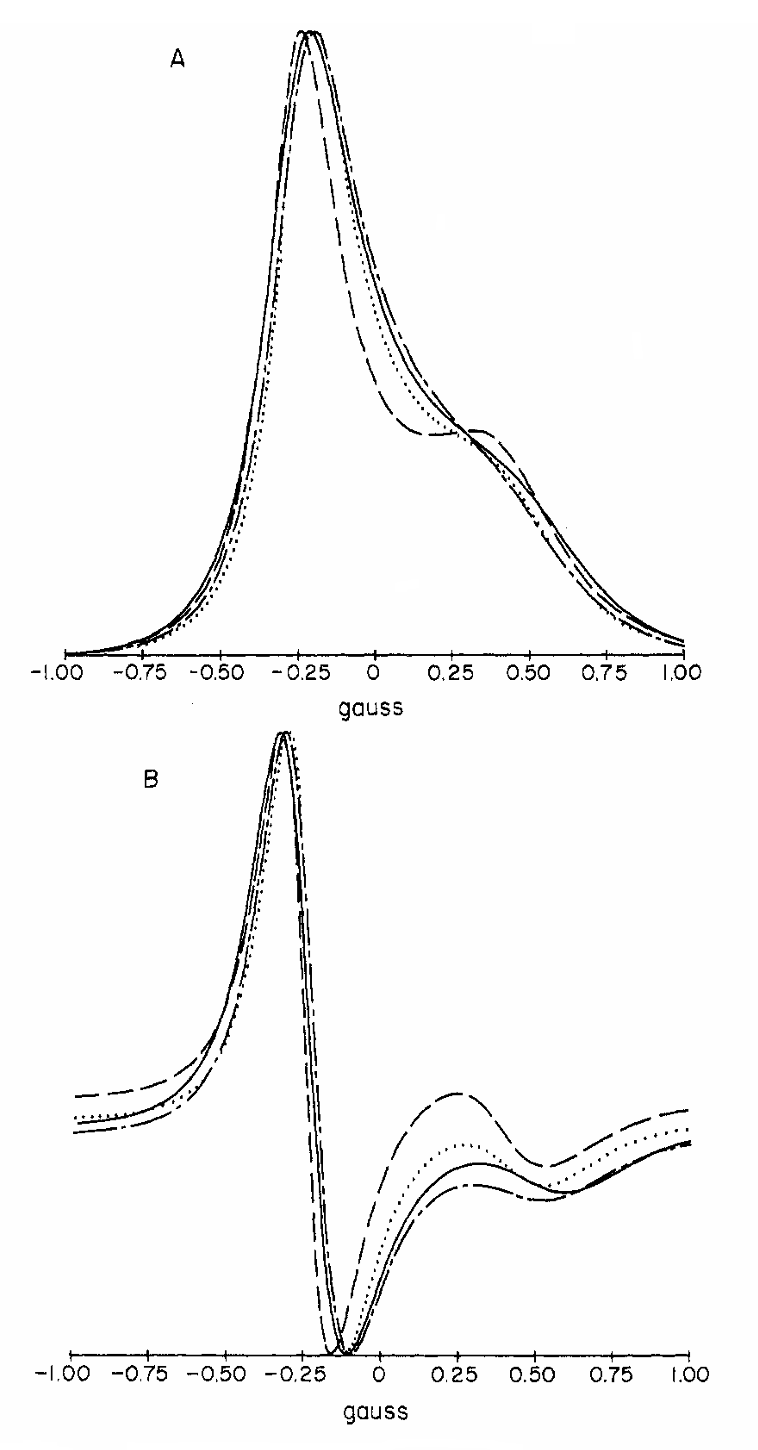
|
|
ABSTRACT: An analysis is given of the inertial effects (which arise from the coupling of the molecular orientational degrees of freedom to the molecular angular momentum) on esr spectra of radicals in the model-sensitive slow tumbling region. The analysis is based on the stochastic-Liouville equation in combined spin, orientational, and angular momentum space, and it utilizes recently developed models of Langevin (or Brownian) diffusion and extended diffusion. The case of a simple line from an axial g tensor is studied in most detail. It is shown that the complete analysis for inertial effects in this case yields results similar to a very crude model of free diffusion found by Goldman, et al., to be in good agreement with experimental results in their analysis of slow-tumbling nitroxide spectra. Results of Langevin and extended diffusion were virtually the same. However, results obtained for a familiar approximate inertial model, which neglects noncommutativity of the angular-velocity components, disagreed sharply with all other cases (including the even cruder free diffusion model derived from it) indicating that it is an unsatisfactory model. The less complete results obtained for the case of nitroxide spectra indicate similar conclusions.
|
|
|
Estimating Microsecond Rotational Correlation Times from Lifetime Broadening of Nitroxide Electron Spin Resonance Spectra Near the Rigid Limit
R.P. Mason and J.H. Freed.
J. Phys. Chem. 78, 1321-1323 (1974).
<doi: 10.1021/j100606a015>
Publication #44
|
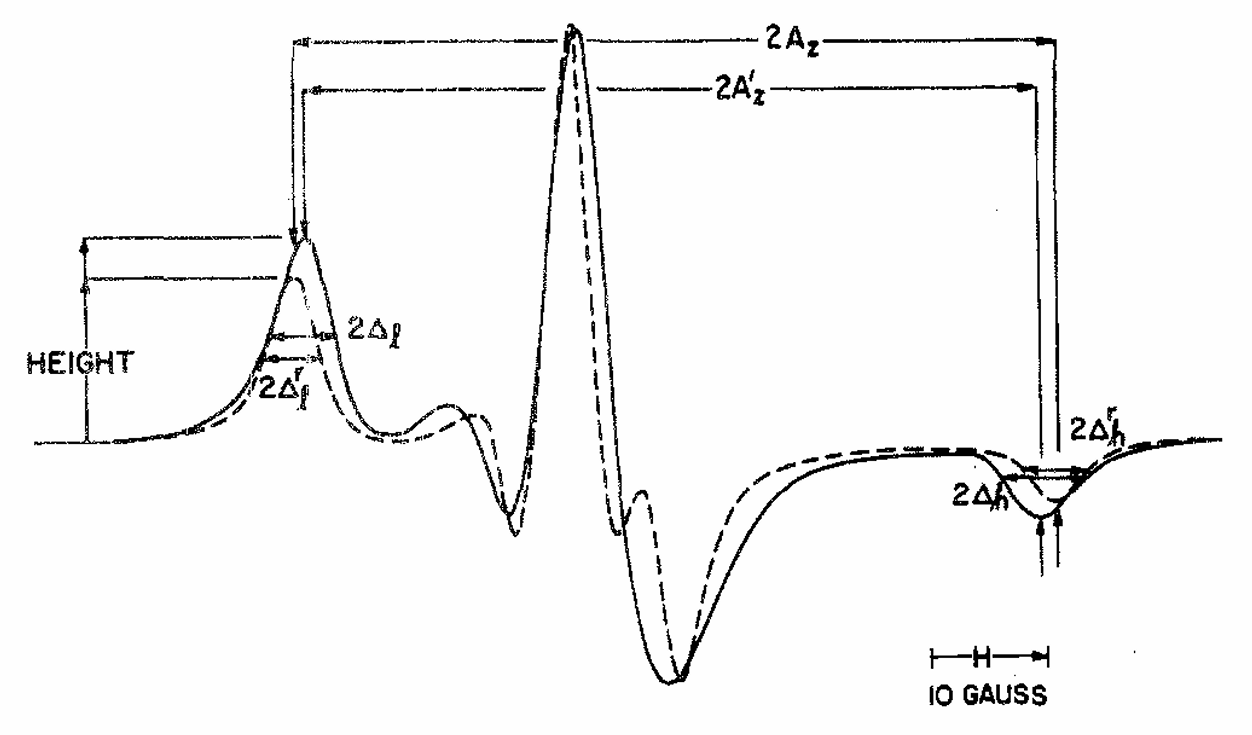
|
|
ABSTRACT: A simple method of estimating rotational correlation times (τR) of the order of microseconds using the widths of the outer esr hyperfine extrema is discussed. An earlier method allowed determination of τR from the ratio of the outer hyperfine extrema separation at a given τR to the rigid limit separation. The present method, however, permits the determination of τR even when the separation of the outer hyperfine extrema is experimentally indistinguishable from the rigid limit value. It should be useful for many macromolecules with a rigidly bound nitroxide spin label, since they typically exhibit values of τR on the order of microseconds in solution.
|
|
|
ESR Studies of Anisotropic Rotational Reorientation and Slow Tumbling in Liquid and Frozen Media. II. Saturation and Nonsecular Effects
S.A. Goldman, G.V. Bruno, and J.H. Freed.
J. Chem. Phys. 59, 3071-3091 (1973).
<doi: 10.1063/1.1680446>
Publication #43
|
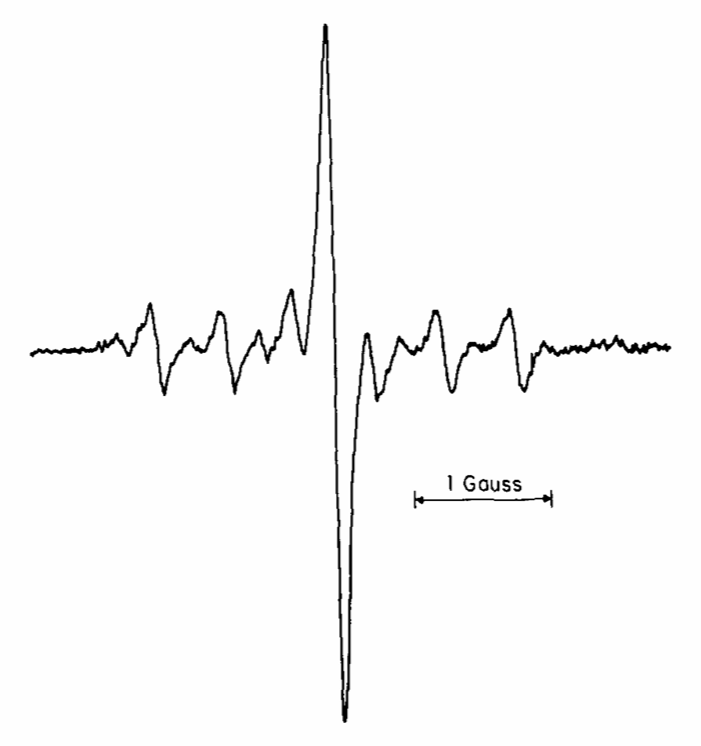
|
|
ABSTRACT: Careful studies are described of the ESR line shapes and saturation behavior for the peroxylamine disulfonate (PADS) radical dissolved in a range of glycerol-water mixtures. This permitted studies where the rotational correlation time τR ranged from 3 × 10−12 sec to < 10−6 sec. The unsaturated line shapes for 10−10 < τR < 10−9 sec are, as in Paper I, readily interpreted in terms of anisotropic rotational diffusion, but for τR < 10−10 sec anomalous behavior of the linewidths, which could be attributed to nonsecular spectral densities, occurs. Supporting experiments at 35 GHz, and also on 17O-labeled PADS, are in excellent agreement with values of τR and N = 4.7 (the ratio of the two components of the axially symmetric diffusion tensor), and they supply further information on the anomalous behavior of the nonsecular spectral densities. A phenomenological treatment based on the memory function approach was found to qualitatively reproduce some observed features. The saturation studies were performed over both the motional narrowing and slow-tumbling regions. The motional narrowing results could be analyzed in a straightforward manner to yield values of We, the electron-spin flip rate, which are found to exhibit a weak dependence on τR, roughly as τR−(1/4) for the glycerol solvents. The stochastic Liouville method is applied to an analysis of slow-tumbling saturated spectra and generally reasonable agreement with experiment is achieved for the simplifying assumptions utilized. The rotationally invariant We obtained from the slow-tumbling analyses are found to agree with the values of We extrapolated from the motional-narrowing region. Other aspects of the slow-tumbling saturation experiments and analysis are discussed.
|
|
|
Theory of Chemically Induced Dynamic Electron Polarization. II
J.B. Pedersen and J.H. Freed.
J. Chem. Phys. 59, 2869-2885 (1973).
<doi: 10.1063/1.1680418>
Publication #42
|
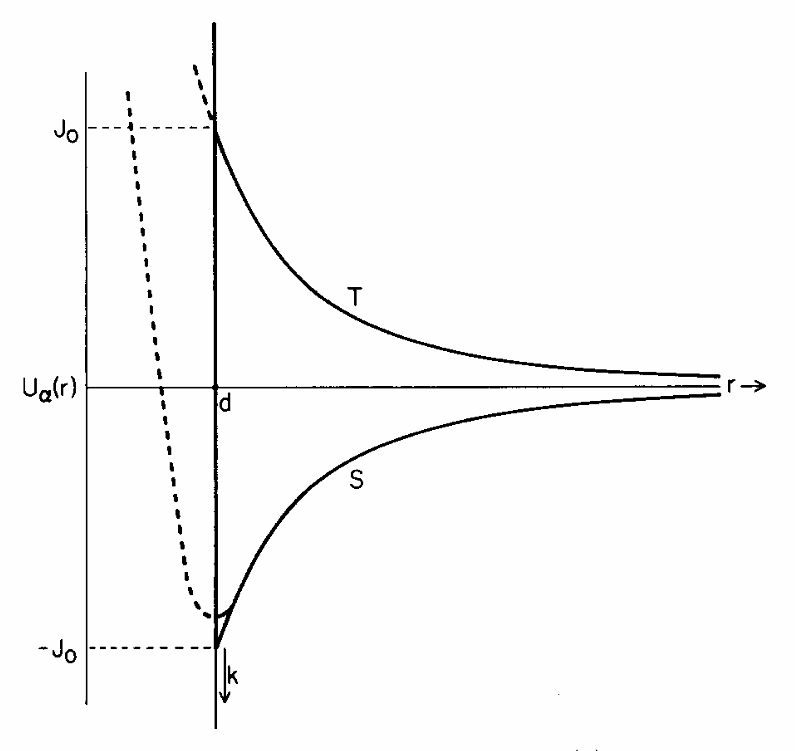
|
|
ABSTRACT: The earlier theoretical analysis for chemically induced dynamic electron polarization (CIDEP), based on the stochastic-Liouville equation, is generalized to explicitly include the spin-dependent exchange forces in the diffusive trajectories, thus permitting a consistent analysis of the simultaneous effects of exchange on both the spin-selective chemical reaction and CIDEP effects. The semiclassical treatment of diffusion under a "classical" force field due to the valence interactions requires the introduction of spin-dependent diffusive and reactive trajectories, and this is discussed for the Brownian-motion model utilized. Our results show that the polarization generated per fractional probability that singlets react (P∞∕ℱ) , is not sensitive to the actual details of the spin-selective reactive process (although the absolute polarization P∞ is sensitive to the reactive process), due presumably to the spatial distinction between interradical separations (r) for which the reaction may occur vs those for which CIDEP polarizations are developed. The former require ℏ ∣ J(r) ∣ ∕ kT > 1 while for the latter ℏ ∣ J(r) ∣ ∕ kT < 1, where J(r) is the exchange interaction. It is found that differences in the (nonreactive) diffusive trajectories for singlets and triplets give polarizations that are generally negligible compared to those which develop as a result of the spin-selective reaction (for our overdamped diffusive model). However, our results for more long-range Coulomb interactions between charged radicals show they can produce significant changes on P∞∕ℱ that are quite sensitive to the magnitude of J. Thus ionic-concentration effects on
should be an important indicator of the CIDEP mechanism. Results are also given for the spin-depolarization process, whereby the effects of spin exchange on a radical pair, which initially collide with residual nonthermal polarization, are to destroy this polarization. The effective range of the spin exchange is found to be weakly enhanced as the range of J(r) is increased. Also, it is shown that, for several variations of a simple exponential dependence of J(r) on r, P∞∕ℱ is hardly affected, although nonexponential dependences can introduce marked changes.
|
|
|
ESR Line Shapes in the Slow-Motional Region: Anisotropic Liquids
C.F. Polnaszek, G.V. Bruno, and J.H. Freed.
J. Chem. Phys. 58, 3185-3199 (1973).
<doi: 10.1063/1.1679640>
Publication #41
|
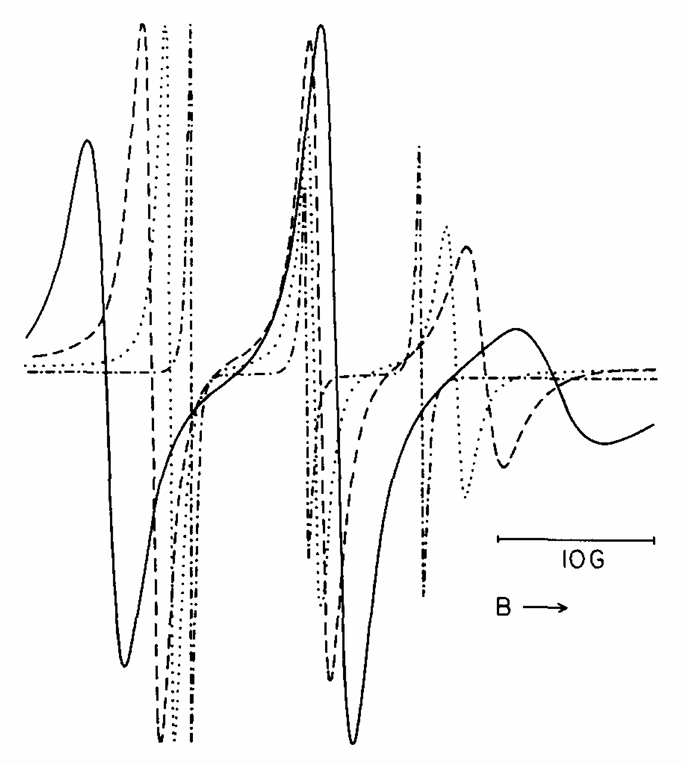
|
|
ABSTRACT: It is shown how the analysis of Freed et al. for ESR lineshapes in the slow tumbling region may be generalized to include anisotropic liquids. Particular emphasis is given to the case of nitroxide radicals in liquid crystal environments with cylindrically symmetric restoring potentials U(β). It is found that when ∣ U(β) ∣ ≲ kT, spectral appearances are qualitatively (but not quantitatively) similar to those for isotropic liquids. In particular, the spectra are sensitive to the model of reorientation. They are also predicted to be a very sensitive indicator of effects of anisotropic viscosity. The analysis given for the motional narrowing region yields analytic expressions for the needed spectral densities, where previously only numerical results had been obtained. The analytic expressions are valid when ∣ U(β) ∣ ≲ kT. Analytic solutions to the rotational diffusion equation appropriate for ∣ U(β) ∣ ≫ kT are given and it is outlined how they may be applied to magnetic resonance.
|
|
|
Theory of Chemically Induced Dynamic Electron Polarization. I
J.B. Pedersen and J.H. Freed.
J. Chem. Phys. 58, 2746-2762 (1973).
<doi: 10.1063/1.1679576>
Publication #40
|
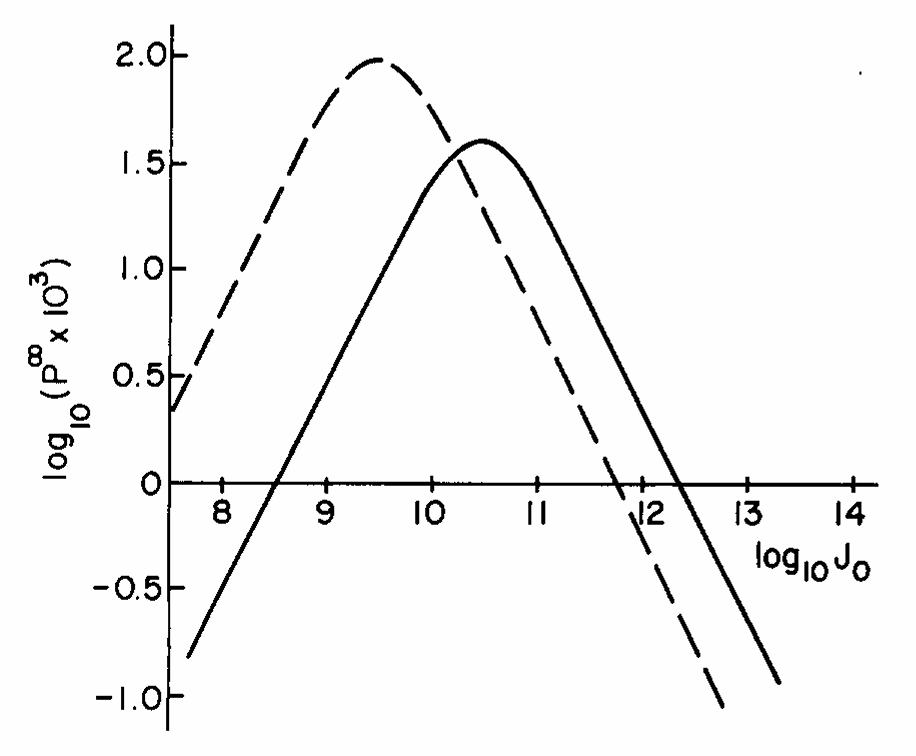
|
|
ABSTRACT: A general and detailed analysis is given of the phenomenon of chemically-induced dynamic electron polarization (CIDEP) by means of the stochastic-Liouville method in accordance with the earlier preliminary report. The finite-difference technique employed permits rapid and convergent solutions without requiring any untoward assumptions on the nature of the models. The dependence of the polarization on the exchange interaction J(r), the Larmor frequency differences between the interacting pair of radicals, diffusion rates, and rates of spin-selective chemical reactions are given in detail. It is shown that models in which J(r) is taken to decay exponentially with r, the radical-separation distance of the radical pair, yield results which are distinctly different from those for a contact exchange model, when J0 [the value of J(r) when r is at the distance of closest approach] is appreciable. The former, more realistic model yields substantial polarizations asymptotically independent of J0, but larger the slower the decrease of J(r) with r; the contact exchange model, however, rapidly goes to zero with increasing J0. These asymptotic values of polarization are predicted to be as high as 10–40 times the equilibrium polarizations (Peq) for sensible values of the relevant parameters, while for values of J0 yielding maximum polarizations (generated at the formative reaction), they can be greater than 100 Peq. These results are of the correct order for agreement with recent experiments. The polarizations have been related to the CIDEP intensities that one may observe for typical schemes of radical production, reaction, and relaxation in order to allow a comparison of the theoretical predictions with experiment.
|
|
|
Theory of Saturation and Double Resonance in ESR Spectra. V. The Average ENDOR and ELDOR Lines
J.H. Freed, D.S. Leniart, and H.D. Connor.
J. Chem. Phys. 58, 3089-3105 (1973).
<doi: 10.1063/1.1679620>
Publication #39
|
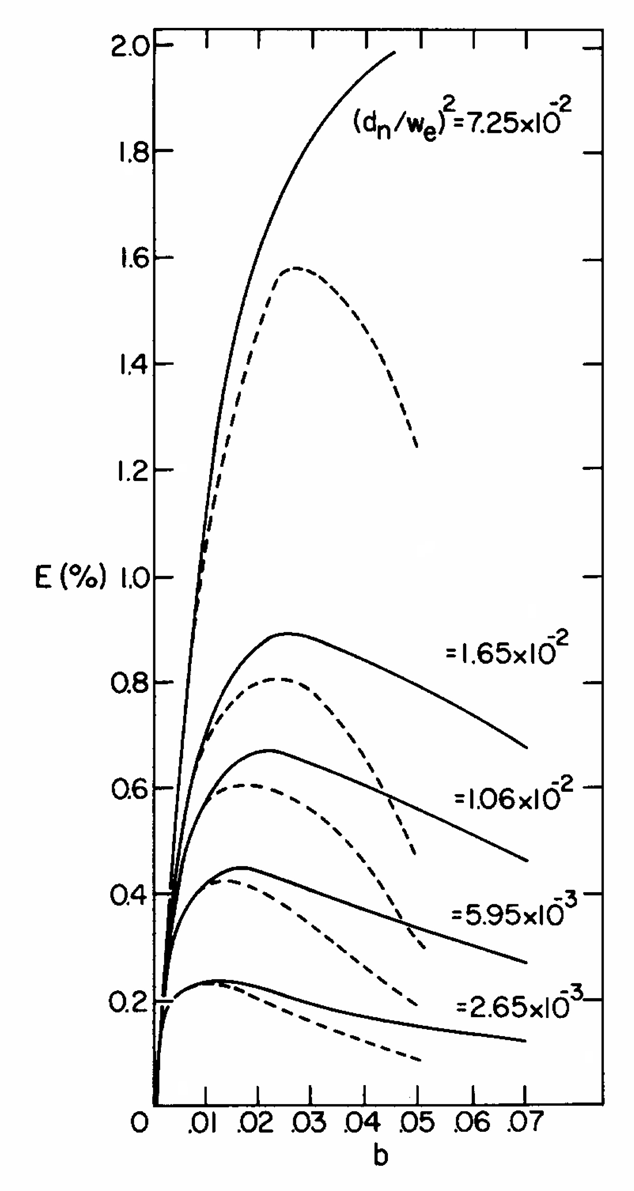
|
|
ABSTRACT: The general theory for the analysis of saturation and double resonance effects in the ESR spectra of dilute solutions of free radicals developed in the earlier papers in this series, has been solved approximately to obtain relatively simple formulas for the general case of free radicals with many nuclei. The solution is based on an expansion linear in b ≡ Wn ∕ We (where Wn and We are, respectively, the lattice-induced pure nuclear-spin and electron-spin flip relaxation rates), but it is asymptotically valid for Heisenberg and (polarized) chemical exchange processes. Also, coherence effects are neglected. The resulting expressions for ENDOR take the simple form of a single average (saturated) Lorentzian for each distinct ENDOR line obtained from a particular group of equivalent nuclei; while for ELDOR one similarly obtains an average ELDOR line shape. Expressions are given for the average saturation parameters that are needed in terms of the various relaxation mechanisms. The range of validity of these solutions is analyzed and discussed. In the case of equivalent nuclei of spins of 1∕2, an interesting result is that the relative ENDOR enhancement of the ESR signal is in lowest order, approximately directly proportional to the number of equivalent spins (I = 1∕2) under certain conditions such as when the NMR resonance is not significantly saturated. This result, when applicable, can prove quite useful for analytical purposes. It is, however, no longer valid for ENDOR signals from groups of equivalent nuclei for which I > 1∕2, and it must be modified, even for I = 1∕2, if lattice-induced cross transitions involving combined electronic and nuclear spin transitions are more important than pure Wn, and these considerations are discussed. Effects of having equivalent nuclei which are not completely equivalent are also discussed. Details regarding the structure and simplifying symmetry considerations of the general solutions are also given.
|
|
|
Calculation of Magnitudes of Chemically Induced Dynamic Electron Polarizations
J.B. Pedersen and J.H. Freed.
J. Chem. Phys. 57, 1004-1006 (1972).
<doi: 10.1063/1.1678279>
Publication #38
|
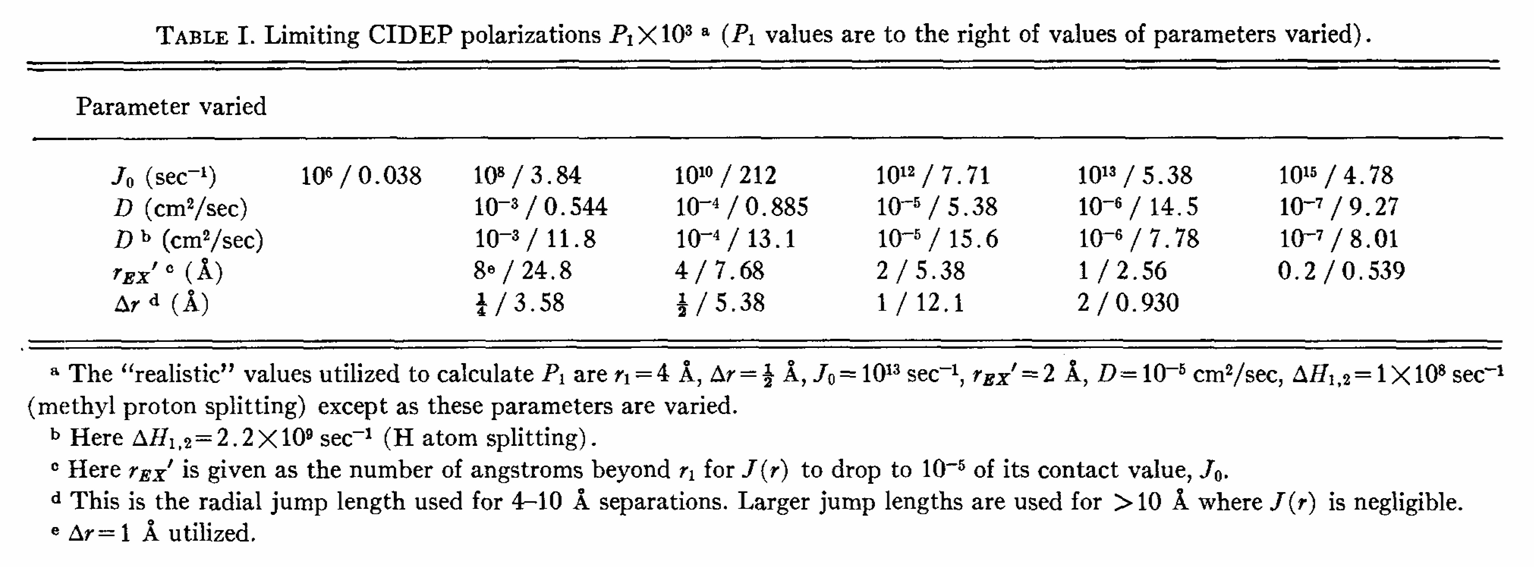
|
|
INTRODUCTION: Recently, there have been several very interesting observations of anomalous ESR spectra arising from nonequilibrium populations of the spin levels for some short-lived free radicals generated in liquids by irradiation or chemical reaction. There have been several initial attempts to explain such observations (in a manner analogous to those offered for CIDNP) in terms of the combined effects of (1) an exchange interaction between a radical pair formed from a dissociating molecule and (2) differences in the Larmour frequencies between the two radicals in the pair by virtue of different g values or different hyperfine interactions with their respective magnetic nuclei. Perhaps the most satisfying analysis to date is that given by Adrian, who considered the situation where the radical pair may separate by diffusion, but then suffers a new encounter. This has the particular advantage of allowing the polarization process to be broken up into two steps of differing time duration. Adrian's approach, while highly instructive, is still incomplete in that he is not able to calculate average quantities involving averaging of the effects of the exchange interaction over the range of possible radical-pair trajectories; he could only guess at order-of-magnitude values.
|
|
|
Electron Spin Resonance
J.H. Freed.
Annu. Rev. Phys. Chem. 23, 265-310 (1972).
<doi: 10.1146/annurev.pc.23.100172.001405>
Publication #37
|
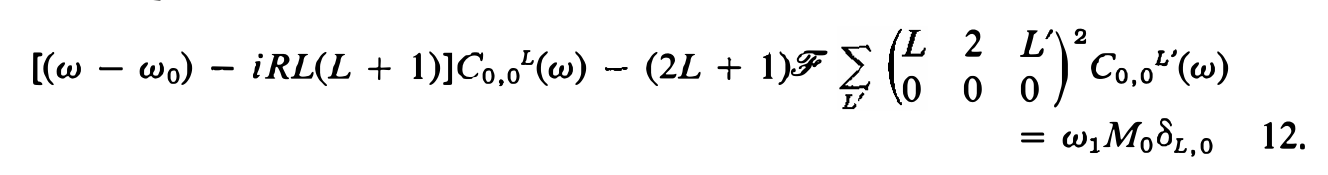
|
|
ABSTRACT: Symons, in the last review of ESR in the Annual Review of Physical Chemistry, has already noted that the vast applications of ESR in the fields of chemistry, physics, and biology render it virtually impossible to attempt anything like a full coverage of the literature. I have, therefore, also adopted the approach of selecting topics that are primarily of interest to me, but have found that space and time limitations have made it necessary to reduce even these.
The main coverage given may be roughly described as the broad overlap region between the ESR of liquids and spin-relaxation phenomena. However, other topics directly or indirectly related (sometimes perhaps only by the author's imagination) are also included. I have emphasized more recent developments and some recent theoretical developments, including the new theoretical and experimental results on ESR spectra for slow tumbling radicals, rotational reorientation in liquids and solids, spin relaxation theory and line width studies, ion pairs, the new phenomenon of chemically induced electron polarization (CIDEP), and the (probably) related old one of Heisenberg spin exchange, saturation and double resonance studies, liquid crystals, and finally gas phase ESR. The most recent detailed review of ESR relaxation in liquids has been given by Hudson & Luckhurst, who covered the literature through 1967. The literature surveyed here covers primarily the period 1969 through 1971. Virtually no coverage has been given to the important applications in structural studies and hyperfine interactions, triplet states and triplet dynamics, radicals in (and adsorbed on) solids, and biological studies.
Part of the problem of dealing with the vast and growing literature in magnetic resonance may be alleviated by the appearance of a new quarterly journal Magnetic Resonance Review, which will survey this literature. In particular, Bolton has already covered the literature of 1970 for ESR in solution and it will continue to be done.
Recent international activities in ESR include the second symposium on ESR spectroscopy at the University of Georgia, December 7-9, 1970. NATO Summer School Lectures on ESR Relaxation in Liquids held in Norway in 1971 are to appear in book form and are frequently referred to in this review.
A most important publication has been the book by Abragam & Bleaney, which provides very extensive coverage of ESR of transition ions and discussions of many general aspects of ESR. Poole & Farach have written a book which reviews relaxation in magnetic resonance, and electron-spin relaxation in solids has been discussed by Standley & Vaughan. Recent introductory texts on ESR include the ample coverage by Bolton & Wertz and a briefer one by Gerson. Discussions and reviews of the biological applications of ESR are given in several places. A review covering, in part, dynamical aspects of triplet state spectroscopy has appeared in last year's Annual Review of Physical Chemistry. A review of halogen hyperfine interactions has been given by Hudson & Root. Other reviews are referred to in the discussions of the relevant topics below.
|
|
|
Estimating Slow-Motional Rotational Correlation Times for Nitroxides by Electron Spin Resonance
S.A. Goldman, G.V. Bruno, and J.H. Freed.
J. Phys. Chem. 76, 1858-1860 (1972).
<doi: 10.1021/j100657a013>
Publication #36
|
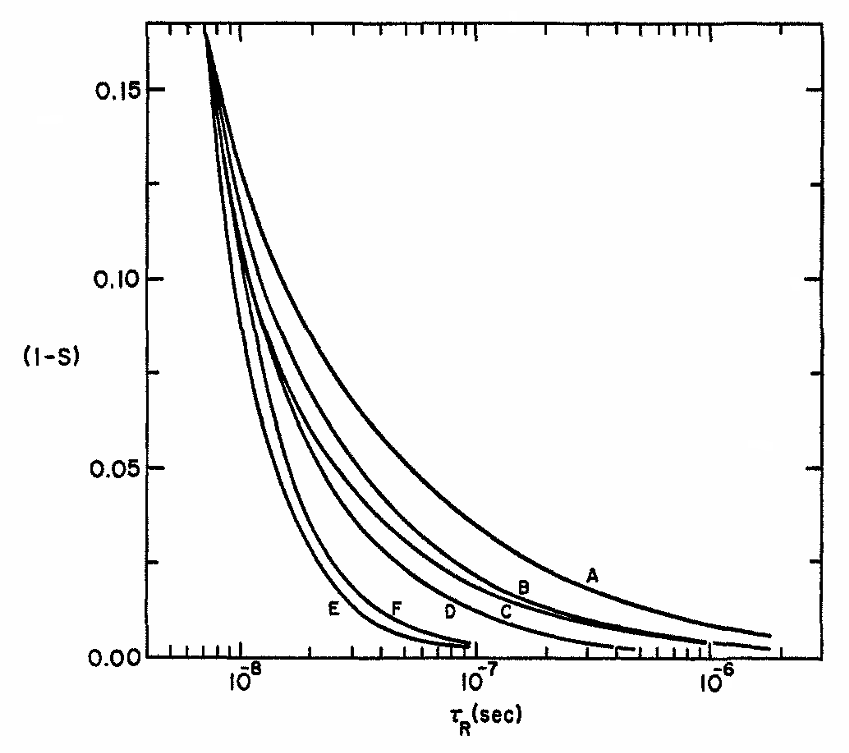
|
|
ABSTRACT: A simple method of estimating slow-motional rotational correlation times τR for nitroxides by esr, which is based on the rigorous theory of Freed, Bruno, and Polnaszek, is discussed. The results can be fit to the expression τR = a(1 − S)b, where S is the ratio of the separation of the outer hyperfine extrema to that for the rigid limit value. The parameters a and b depend upon intrinsic line width, rotational model, and hyper-fine parameters, and appropriate results are given.
|
|
|
Spin—Rotational Relaxation in One Dimension: Angular Momentum-Orientational Correlation
J.H. Freed.
J. Chem. Phys. 56, 1407-1408 (1972).
<doi: 10.1063/1.1677374>
Publication #35
|
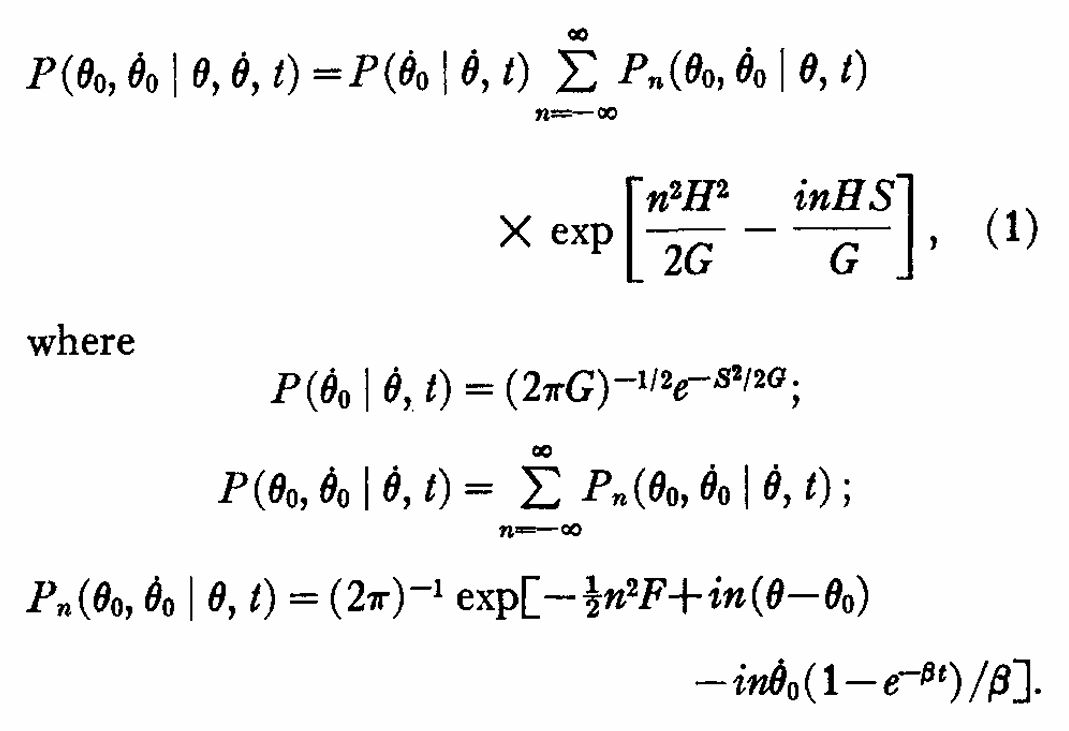
|
|
ABSTRACT: Spin-rotational relaxation is generally accepted as an important spin-relaxation mechanism for both NMR and ESR in liquids. One usually uses the Hubbard model which employs the rotational analog of the Langevin equation, for the calculation of angular momentum correlation functions. That is, one examines the random time-dependent behavior of the spin-rotation interaction term (in units of ℏ): ℋSR = J⋅C⋅S where the molecular angular momentum J and the orientation-dependent spin-rotation tensor C are governed by rotational Brownian motion. In spherical tensor notation, ℋSR = C(0) J⋅S+∑mC (2,m) (SJ)(2,−m). In general one must calculate the correlation function ❬ ℋSR(t) ℋSR*(t+τ) ❭ to obtain spin-relaxation effects. One then finds that for the term involving the spherically symmetric part of C or C(0) = ⅓ TrC, the correlation function ❬ J1(t) Jm(t+r) ❭ is needed, and it decays exponentially with time constant τJ = β−1 = I∕β′, where β′ is the friction constant. However, for the orientation dependent portion of ℋSR one has to consider correlation functions of type: ❬ [ Jm′′ 𝔇0m′L (Ω) ]t+r[ Jk′′ 𝔇0k′(L′) (Ω) ]t ❭ i.e., one needs the joint correlation function for angular momentum and orientation. Since the six dimensional Markoffian conditional probability function in angular velocity and orientation is not known, the usual procedure is to treat the momentum and orientation parts as though they are independent. One then usually relies on the fact that, for Brownian motion, the orientational correlation time τθ,L ≫ τJ, to justify this.
|
|
|
Spin Relaxation via Quantum Molecular Systems
J.H. Freed.
In Electron Spin Relaxation in Liquids, L.T. Muus and P.W. Atkins, Eds.
Plenum Publishing Co.: New York, 1972; Chapter 9, pp. 193-212.
<doi: 10.1007/978-1-4615-8678-4_9>
Publication #34
|
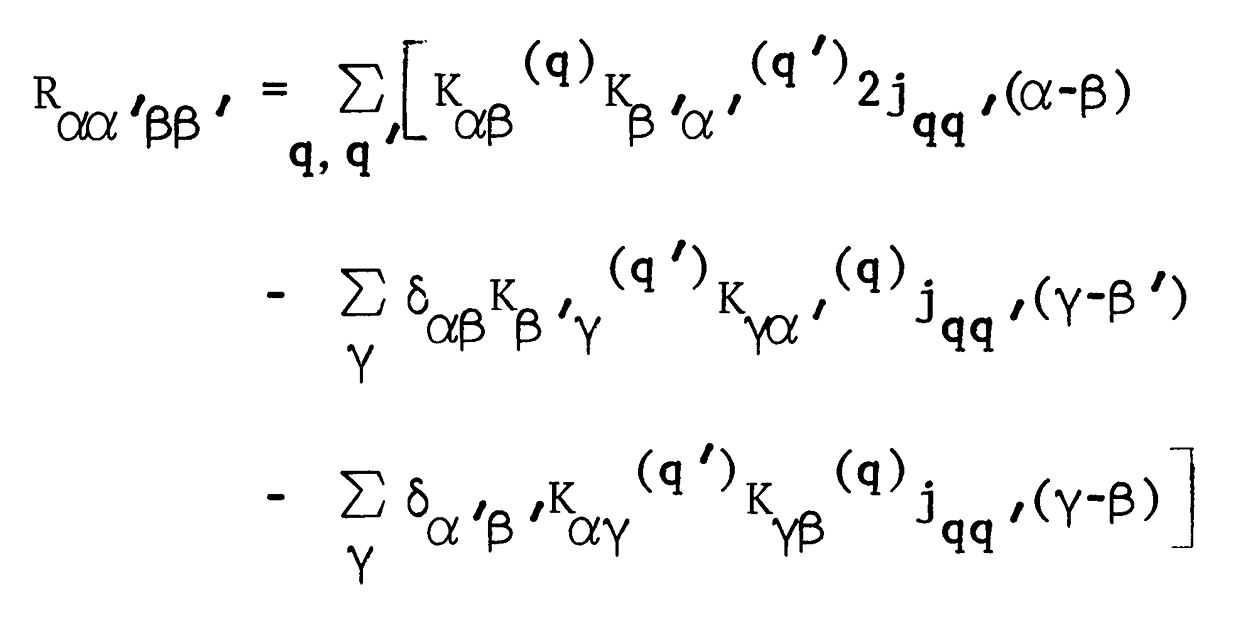
|
|
ABSTRACT: Very often one has to consider the quantum nature of the molecular systems whose modulation induces spin relaxation. We first consider a "gas-like" model wherein strong collisions randomize the molecular degrees of freedom, more specifically the rotational states. Then we generalize the results to cover more general descriptions of the way that the molecular degrees of freedom relax through thermal contact.
|
|
|
|
|
|
|
|
|
ESR Study of Heisenberg Spin Exchange in a Binary Liquid Solution near the Critical Point
J.C. Lang, Jr. and J.H. Freed.
J. Chem. Phys. 56, 4103-4114 (1972).
<doi: 10.1063/1.1677822>
Publication #33
|
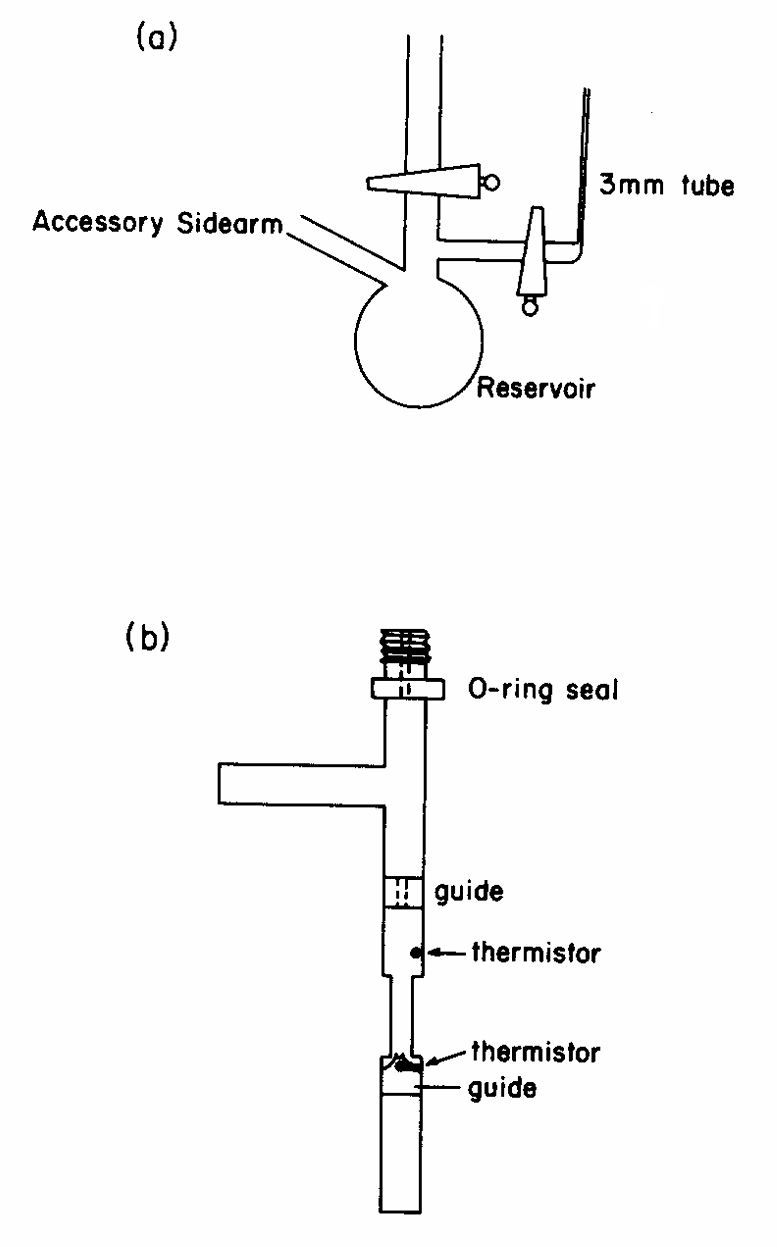
|
|
ABSTRACT: A careful study of the Heisenberg spin-exchange contribution ωHE to the ESR linewidths of the di-t-butyl nitroxide (DTBN) radical dissolved in mixtures of 2,2,4-trimethylpentane and n-perfluoroheptane was performed. The study includes samples of different radical concentration dissolved in the critical composition of the two solvents. This critical solvent system is known to exhibit an anomaly in the macroscopic kinematic viscosity v near Tc. It is found that in the critical region, ωHE is not linear in T∕v. However, it was observed that ωHE is linear in T∕v′ both for noncritical compositions and critical compositions above Tc. Here v′ is the macroscopically measured viscosity, but with the "anomalous portion" subtracted out. Deviations from ideal behavior of ωHE with respect to T∕v were observed and discussed. The experiments near the critical region required temperature stability and control to within ±0.01°C at the ESR sample, and a description is given of the experimental design.
|
|
|
An ESR Study of Anisotropic Rotational Reorientation and Slow Tumbling in Liquid and Frozen Media
S.A. Goldman, G.V. Bruno, C.F. Polnaszek, and J.H. Freed.
J. Chem. Phys. 56, 716-735 (1972).
<doi: 10.1063/1.1677222>
Publication #32
|
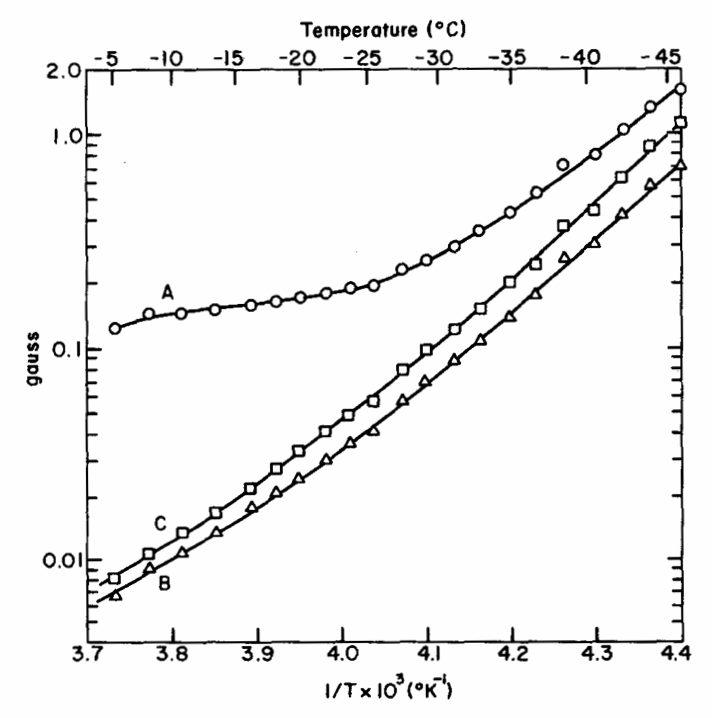
|
|
ABSTRACT: A careful study is described of the ESR lineshapes for the peroxylamine disulfonate (PADS) radical dissolved in 85% glycerol solution and in frozen water and D2O. In the frozen media, spectra characteristic of rotational correlation times τR ranging from 1.0 × 10−11 sec to > 10−6 are obtained, while the range in glycerol is from
to 1 × 10−10 sec to > 10−6. The very rapid rotational motion in frozen water is taken to imply that PADS is rotating in a clathrate cage. The activation energies in ice and 85% glycerol are 14.7 ± 0.1 ans 11.3 ± 0.1 kcal ∕ mole, respectively, (from motional-narrowing data). The value for ice is very similar to that obtained for other rate processes in ice. The lineshapes for τR ≲ 10−9 sec are analyzed in terms of the familiar spin-relaxation theories valid in the motionally narrowed region. These results are well fitted by the model of axially symmetric rotational diffusion with the symmetry axis in the plane of the N, O, and S atoms and parallel to a line passing through the two S atoms. Diffusion about this axis is found to be 2.9 ± 1 and 4.7 ± 1 times faster for frozen water and glycerol solvent, respectively than about the other axes over a wide range of values of average τR̄. It was possible to obtain these results, because accurate measurements of the g and A tensors for PADS in these media could be made from the well-resolved rigid spectra at X band and 35 GHz; the intrinsic widths in D2O are only about 1.5 G. The spectra in the slow-motional region τR ̄ >10−9 sec were simulated utilizing the slow tumbling formulation of Freed, Bruno, and Polnaszek appropriately generalized to include completely asymmetric g and A tensors. The simulated spectra are found, in general, to be in quite good agreement with experimental observations. The agreement is clearly improved by introducing axially symmetric rotational diffusion, as found for the motional-narrowing region, into the simulations. Spectra are simulated for Brownian rotational diffusion as well as for simplified models of free diffusion, which includes inertial effects, and for diffusion by jumps of substantial angle. Improved agreement with experiment is found with some of these latter models. What appears to be a surprisingly small nonsecular linewidth contribution in the motional-narrowing region is briefly discussed in terms of these models.
|
|
|
ESR Line Shapes for Triplets Undergoing Slow Rotational Reorientation
J.H. Freed, G.V. Bruno, and C. Polnaszek.
J. Chem. Phys. 55, 5270-5281 (1971).
<doi: 10.1063/1.1675667>
Publication #31
|
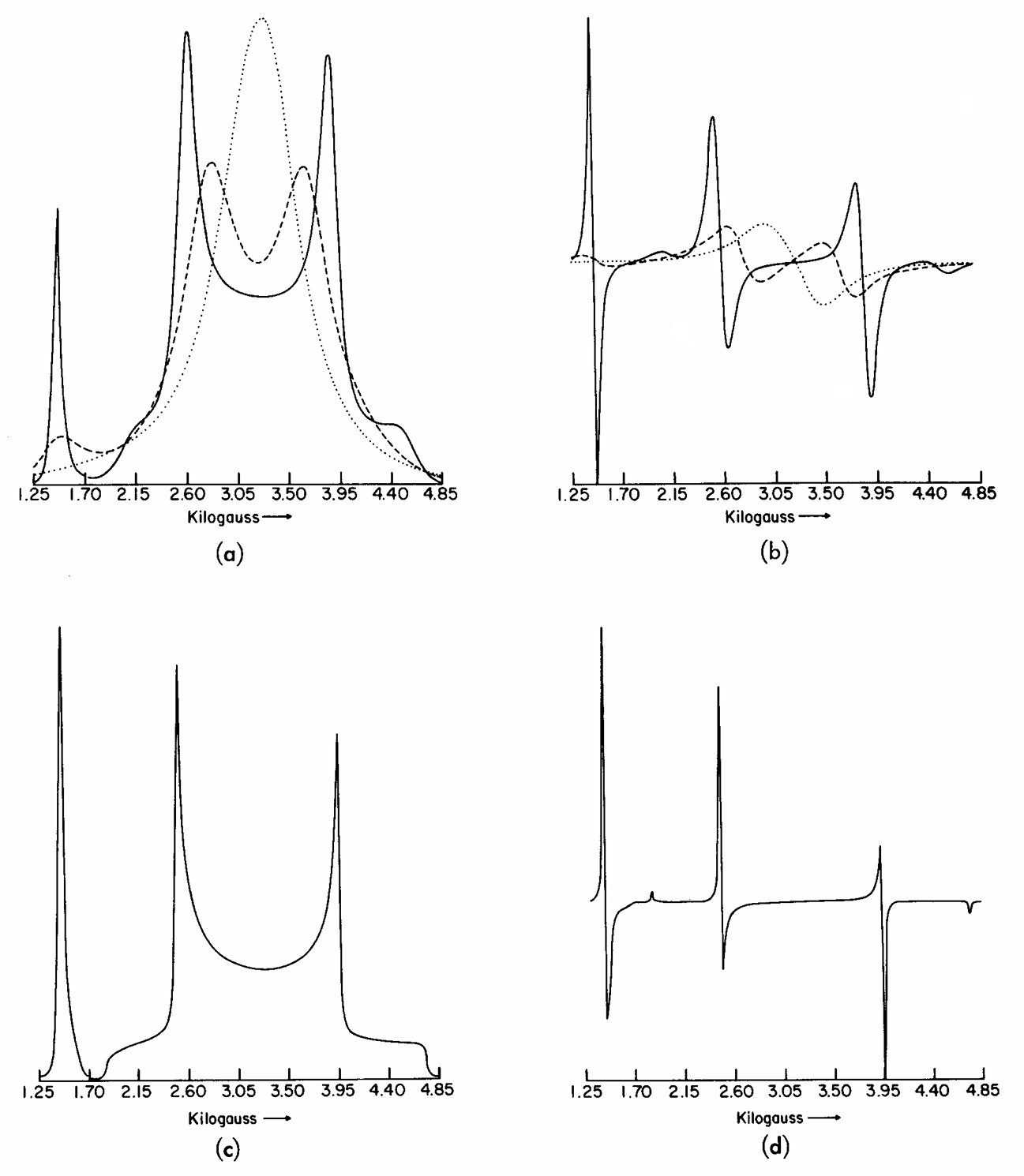
|
|
ABSTRACT: The stochastic Liouville method is applied to analyze the general problem of unsaturated ESR line shapes for triplets undergoing rotational diffusion. Detailed line shape simulations are obtained for the cases of high-, low-, and zero-field resonance, large or small values of an axially symmetric zero-field splitting, and slow through fast rotational diffusion. It is shown how all these parameters can have profound effects on the observed ESR spectra.
|
|
|
Electron Spin Resonance Line Shapes and Saturation in the Slow Motional Region
J.H. Freed, G.V. Bruno, and C.F. Polnaszek.
J. Phys. Chem. 75, 3385-3399 (1971).
<doi: 10.1021/j100691a001>
Publication #30
|
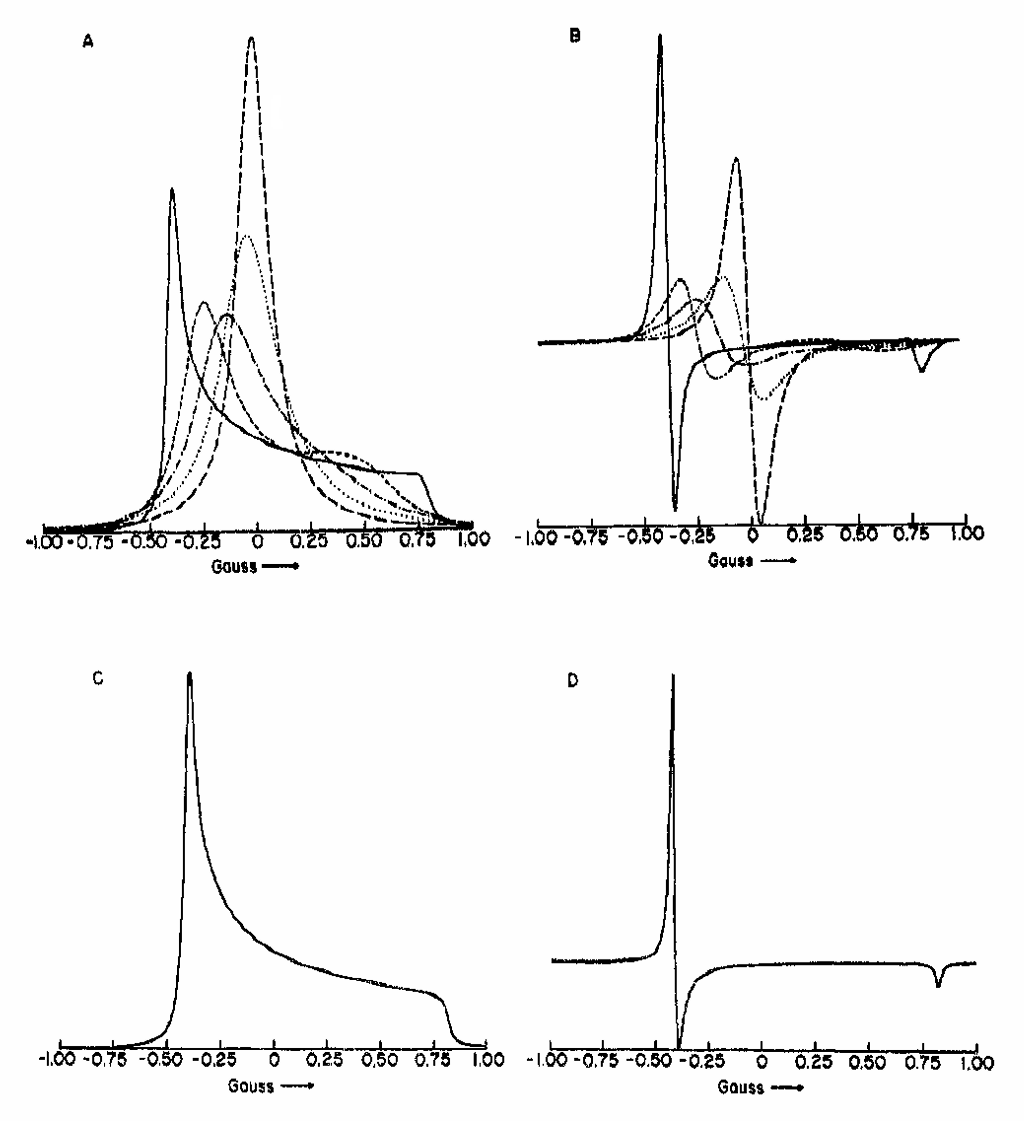
|
|
ABSTRACT: The stochastic Liouville method is developed and applied in a manner appropriate to analyze the problem of esr line shapes and saturation in the slow-motional region with particular emphasis on rotational diffusion. Detailed unsaturated line shape solutions are obtained for axial and asymmetric g tensors and axial dipolar tensors. These line shapes are compared to the predicted rigid-solid spectral shapes. In particular, it is shown that the pseudo-secular dipolar terms make significant contributions to the slow tumbling line shapes expected for 14N-containing radicals such as nitroxides and may not be neglected in such cases. Saturation effects are analyzed for a two-jump model, as well as for rotational diffusion of the g tensor. It is found that even in the slow-motional region, a fundamental role is played by the T1's that are obtained from the spin-relaxation theories. One finds that the significant line shape changes resulting from saturation are dependent on the rotational diffusion rates.
|
|
|
ESR Relaxation Studies on Orbitally Degenerate Free Radicals. II
M.R. Das, S.B. Wagner, and J.H. Freed.
J. Chem. Phys. 52, 5404-5417 (1970).
<doi: 10.1063/1.1672791>
Publication #29
|
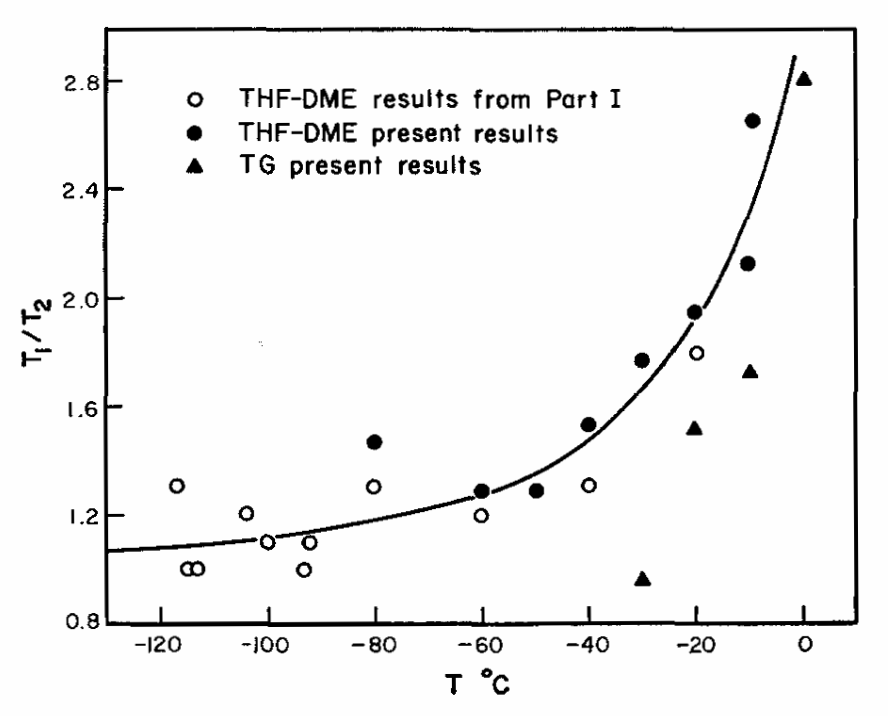
|
|
ABSTRACT: Careful linewidth and saturation studies were performed on several orbitally degenerate hydrocarbon free radicals in liquid solution as a function of temperature, solvent, and counterion, and these are compared with similar studies on nondegenerate hydrocarbon free radicals. The anomalous T1's and T2's attributed to the orbital degeneracy are found to be strikingly independent of all these variables. This is most clearly evidenced for the coronene and triphenylene anion radicals. The results for the benzene and cyclo-octatetraene anions (alkali metal prepared) are complicated by a further contribution to T2−1 which increases with temperature. This contribution is correlated with counterion and solvent dependent ion-pairing effects. The intrinsic temperature-independent T2−1's for the orbitally degenerate free radicals are surprisingly well correlated with the small but anomalous deviations of the g values for these radicals from the Segal, Kaplan, and Fraenkel experimental fit to Stone's theory of g values. This is taken as positive evidence for the role of anomalous spin–orbit interactions in the mechanism(s) yielding the anomalous relaxation times. Their independence of temperature, solvent, and counterion suggests that the relaxation mechanism(s) is largely intramolecular.
|
|
|
An Electron Nuclear Double Resonance and Electron Spin Resonance Study of Semiquinones Related to Vitamins K and E
M.R. Das, H.D. Connor, D.S. Leniart, and J.H. Freed.
J. Am. Chem. Soc. 92, 2258-2268 (1970).
<doi: 10.1021/ja00711a011>
Publication #28
|
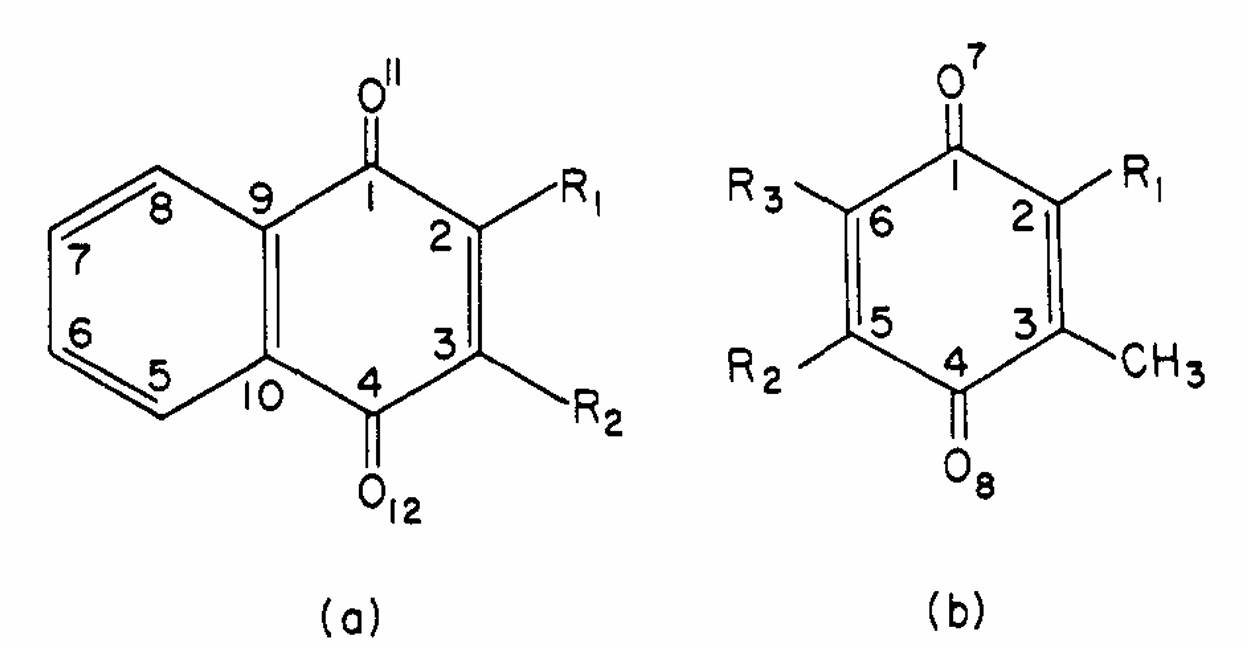
|
|
ABSTRACT: The electron nuclear double resonance (endor) spectra of ethanolic solutions of several semiquinones of biological interest have been observed, and splitting constants as well as per cent enhancement measurements have been made. The endor results, which are especially useful in obtaining unequivocal analyses of the more complex esr spectra, also showed greatly increased resolution by resolving small splitting constant differences unobservable in the esr. This latter result was due to a factor of ˜5 decrease in the endor as compared to the esr widths. These results were used to evaluate spin densities. MO calculations which were performed were found to be in satisfactory agreement with the "experimental" spin densities, including those at positions of small density. It is found that the introduction of a long side chain in the vitamin semiquinones in place of a methyl group has very little effect on the spin density distribution, including that at the ring carbon to which the side chain is attached. Alternating line width effects were observed in the esr of the semiquinones derived from vitamin K1 and coenzyme Q-10 (ubisemiquinone, USQ) with associated line width effects in the endor. They are due to rotation of the aromatic ring relative to the attached methylene protons of the side chain. At -50°, this motion is sufficiently slow in USQ in dimethoxyethane (DME) that two inequivalent methylene protons are observed in the endor and esr. Comparable activation energies and preexponential factors were found for USQ in DME and ethanol with similar results for vitamin K1.
|
|
|
ESR Studies of Heisenberg Spin Exchange. III: An ELDOR Study
M.P. Eastman, G.V. Bruno, and J.H. Freed.
J. Chem. Phys. 52, 321-327 (1970).
<doi: 10.1063/1.1672686>
Publication #27
|
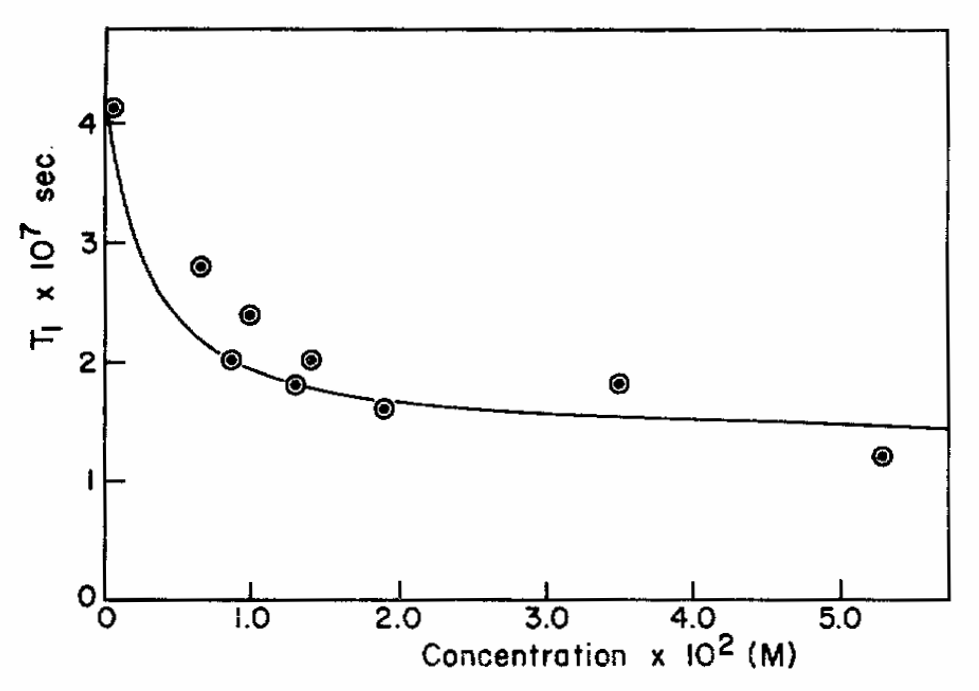
|
|
ABSTRACT: We report the first quantitative test of the theory for electron–electron double resonance (ELDOR) of free radicals in solution. The reduction factors R for the ELDOR spectra of aqueous solutions of the peroxylamine disulfonate dianion radical (PADS) were examined and shown to fit the theory given by Hyde, Chien, and Freed in the extrapolated limit of infinite pump power. Some of the experimental and theoretical problems are discussed. The effect of exchange on the saturation properties of PADS was investigated and shown to agree, within experimental error, with the theory discussed in Part I of this series. The implications of these results for the determination of relaxation times in complex systems are briefly discussed.
|
|
|
ESR Studies of Heisenberg Spin Exchange. II. Effects of Radical Charge and Size
M.P. Eastman, G.V. Bruno, and J.H. Freed.
J. Chem. Phys. 52, 2511-2522 (1970).
<doi: 10.1063/1.1673335>
Publication #26
|
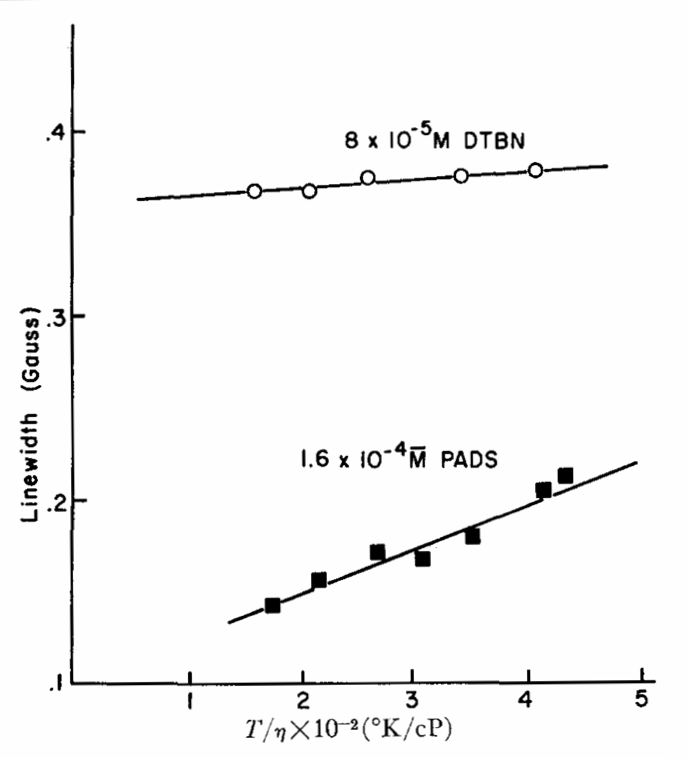
|
|
ABSTRACT: The Heisenberg spin exchange behavior of several radical–solvent systems has been studied. This work has shown that 𝓀, the second-order rate constant for bimolecular encounters, and ∣J∣ τ1 (where ∣J∣ is twice the exchange integral and τ1 is the lifetime of the collision pair) depend markedly on the ionic strength of the solution and upon the size and charge of the radical. For the peroxylamine disulfonate radical in a 5 × 10−2M solution of 1:2 electrolyte, our simple analysis yields ∣J∣ τ1 ˜0.95 and 𝓀 = 2.0 × 10+9M−1 ⋅ sec−1 at 24°C; increasing the electrolyte concentration to 6.1 × 10−1M gives ∣J∣ τ1˜1.3 and 𝓀 = 3.2 × 109M−1 ⋅ sec−1. These changes in 𝓀 and ∣J∣ τ1 are attributed to screening by the electrolyte of the charge of the dianion radical; this is discussed in terms of the Debye theory. The di-tertiarybutyl nitroxide (DTBN) radical undergoes apparent strong exchange (∣J∣ τ1 ≫ 1) in pure water (𝓀 = 2.4 × 109M−1 ⋅ sec−1 at 24°C). The dependence of the exchange frequency (ωHE) on added 1:2 electrolyte is of the form ωHE = ωHE(0) exp(−CE ∕ C′), where CE is the molar electrolyte concentration and ωHE(0) the exchange frequency in pure water; for a 2.4 × 10−2M DTBN solution C′ = 0.23 ± 0.02M. This reduction of exchange frequency with electrolyte concentration is attributed to the formation of DTBN aggregates. The durosemiquinone radical undergoes strong exchange in dimethoxyethane (𝓀 = 2.9 × 109M−1 ⋅ sec−1 at 15 °C). The closeness of 𝓀 to the diffusion-controlled limit indicates that ion pairing takes place in solution. The tetraphenyl arsonium peroxylamine disulfonate radical undergoes apparent weak exchange (∣J∣ τ1 < 1) in dimethyl sulfoxide. However, rapid radical decay made quantitative study of exchange impossible for this radical.
|
|
|
Studies of Heisenberg Spin Exchange in ESR Spectra. I. Linewidth and Saturation Effects
M.P. Eastman, R.G. Kooser, M.R. Das, and J.H. Freed.
J. Chem. Phys. 51, 2690-2709 (1969).
<doi: 10.1063/1.1672395>
Publication #25
|
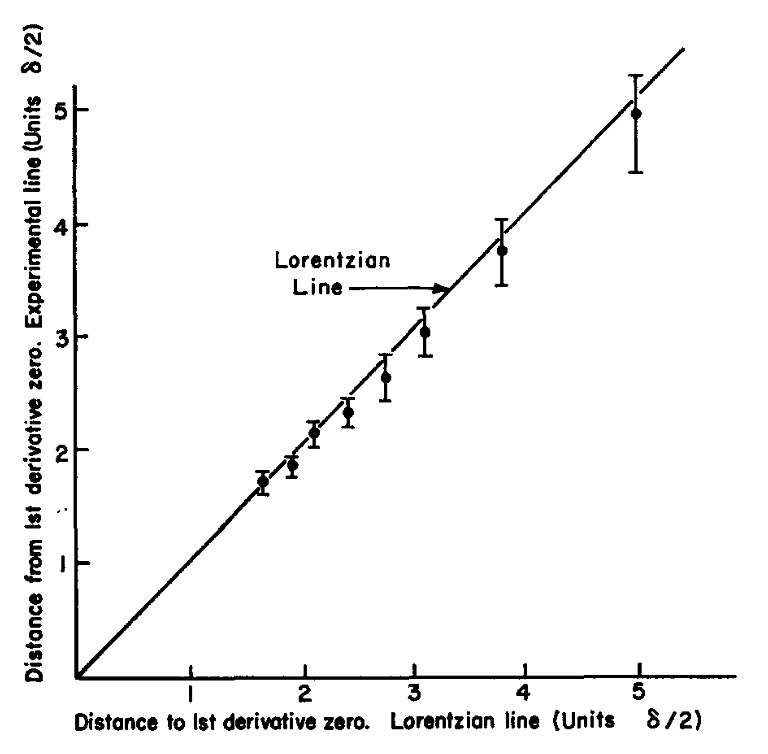
|
|
ABSTRACT: A compact general theory for the effect of Heisenberg spin exchange on ESR linewidths and saturation parameters is detailed. The effects of Heisenberg exchange on the linewidths of the tetracyanoethylene anion (TCNE−) radical and the di-tert-butyl nitroxide (DTBN) radical in both dimethoxyethane (DME) and tetrahydrofuran (THF) are investigated. From comparative studies of linewidth as a function of temperature and of radical concentration, TCNE− in DME is shown to undergo strong exchange with a second-order rate constant of 4.1 ± 0.6 × 109M−1 ⋅ sec−1 at 15°C. The TCNE− radical in THF exhibits an anomalous concentration-dependent linewidth effect when compared to the theory and to the experiments employing DME as the solvent. The uncharged DTBN radical shows similar spin-exchange properties in both solvents. Possible mechanisms for the anomalous linewidth effect are discussed. The effect of spin exchange on the saturation parameters of the TCNE− radical in DME is investigated in detail, and the experimental results are shown to agree, within experimental error, with the theory developed. Electron–nuclear dipolar and electron–electron dipolar relaxation effects are discussed in terms of their (small) contributions to the experimentally determined relaxation times.
|
|
|
ESR Relaxation Studies on Orbitally Degenerate Free Radicals. I. Benzene Anion and Tropenyl
R.G. Kooser, W.V. Volland, and J.H. Freed.
J. Chem. Phys. 50, 5243-5257 (1969).
<doi: 10.1063/1.1671041>
Publication #24
|
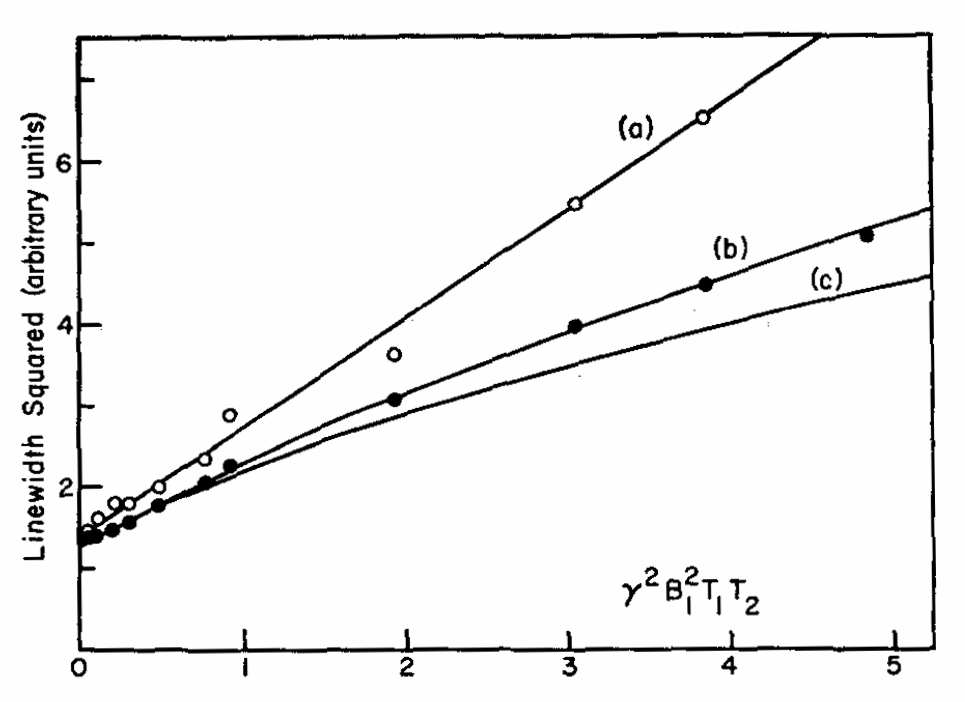
|
|
ABSTRACT: Careful continuous saturation measurements, which include corrections for nonuniform microwave and modulating fields, have been used to examine, at different temperatures, the ESR linewidths and saturation behavior of the benzene anion in a tetrahydrofuran: dimethoxyethane solvent and the tropenyl radical in molten bitropenyl. It has been found that the ratios of the longitudinal to transverse electron-spin relaxation times (T1 ∕ T2) for these free radicals in solution are about 1.1 ± 0.1 for the benzene anion below −60°C and for tropenyl at 165°C. Analysis of the results has shown that at 9.1 GHz, g-tensor, anisotropic dipolar, and spin–rotational relaxation mechanisms do not contribute appreciably to the observed times. The anomalously small relaxation times associated with these radicals have been attributed to effects involving the degenerate ground states of these radicals. The spin-relaxation behavior of peroxylamine disulfonate anion in aqueous solution has also been studied.
|
|
|
Theory of Saturation and Double Resonance Effects in ESR Spectra. IV. Electron-Nuclear Triple Resonance
J.H. Freed.
J. Chem. Phys. 50, 2271-2272 (1969).
<doi: 10.1063/1.1671371>
Publication #23
|
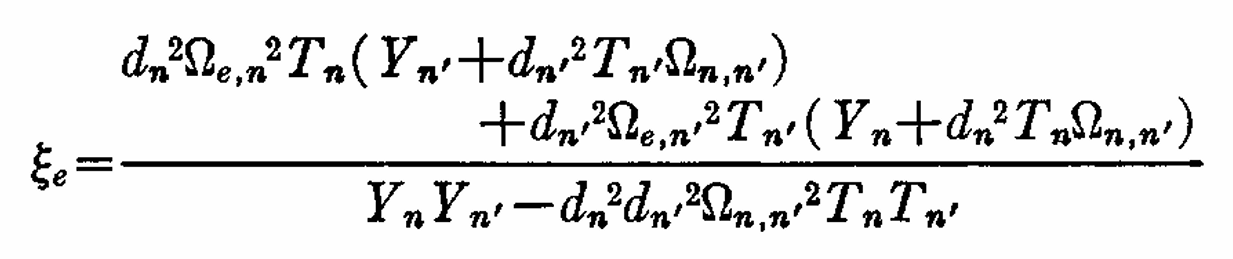
|
|
ABSTRACT: Recent experimental and theoretical studies of the ENDOR technique in liquids have demonstrated its utility in analyzing complex ESR spectra and also its strong dependence on the details of spin relaxation. It was predicted that liquid-state ENDOR enhancements are optimum when the lattice induced nuclear-spin-flip rate (Wn) [usually due to an electron-nuclear dipolar (END) mechanism] is comparable to the lattice-induced electron-spin-flip rate (We). In a simple picture, this is because "shorting out" Wn between the two spin levels b and d makes the relaxation of the saturated a↔b ESR transition occur more rapidly via the alternative path a→c(Wn); c→d(We); d→b (rf induced), as long as Wn and We are still active. In many liquid situations, particularly near room temperature, one expects We ≫ Wn for protons, and a very poor ENDOR enhancement is both predicted and observed. If, however, both nuclear transitions: ωn = | γn B0 ± ½ γe an | corresponding to a particular proton (or set of equivalent protons) are simultaneously saturated, then it should be possible to obtain significant ENDOR enhancements even when We ≫ Wn, because now also the a↔c transition is "shorted out." One could then obtain information equivalent to that of the ENDOR technique, but on a greater variety of samples.
|
|
|
T1/T2 and Spin Relaxation in the Benzene Anion
J.H. Freed and R.G. Kooser.
J. Chem. Phys. 49, 4715-4717 (1968).
<doi: 10.1063/1.1669937>
Publication #22
|
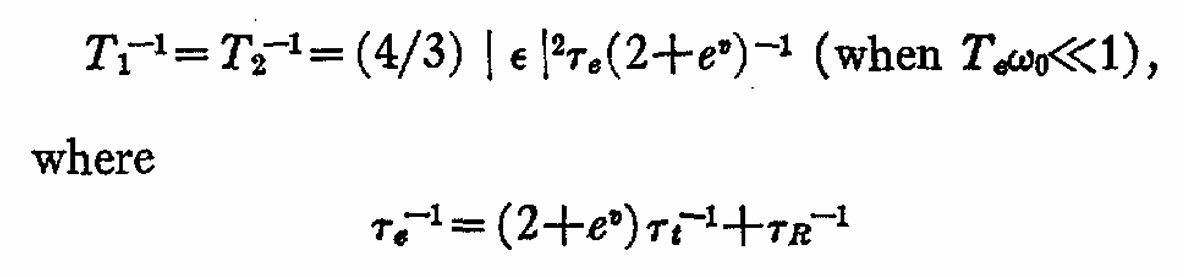
|
|
ABSTRACT: The anomalously broad linewidths of the radical anion of benzene (B−) and associated radicals of high symmetry have been the subject of considerable interest. A number of possible explanations have been proposed which are all based on time-dependent interactions between polar solvent (or cation) and B− that affect the otherwise vibronic ground-state degeneracy of the molecule. There is the crucial question as to which spin-dependent interaction would then be affected, and this can be at least partially answered in terms of comparison with detailed spectral observables. For example, solvent-induced hyperfine fluctuations have been ruled out because of the absence of any significant variation in widths among the hyperfine components. Also, field-dependent contributions were shown not to be important. Spin-orbit (and associated) interactions which are nuclear-spin independent have also been considered in terms of effects peculiar to the case of vibronic degeneracy.
|
|
|
Generalized Cumulant Expansions and Spin-Relaxation Theory
J.H. Freed.
J. Chem. Phys. 49, 376-391 (1968).
<doi: 10.1063/1.1669833>
Publication #21
|
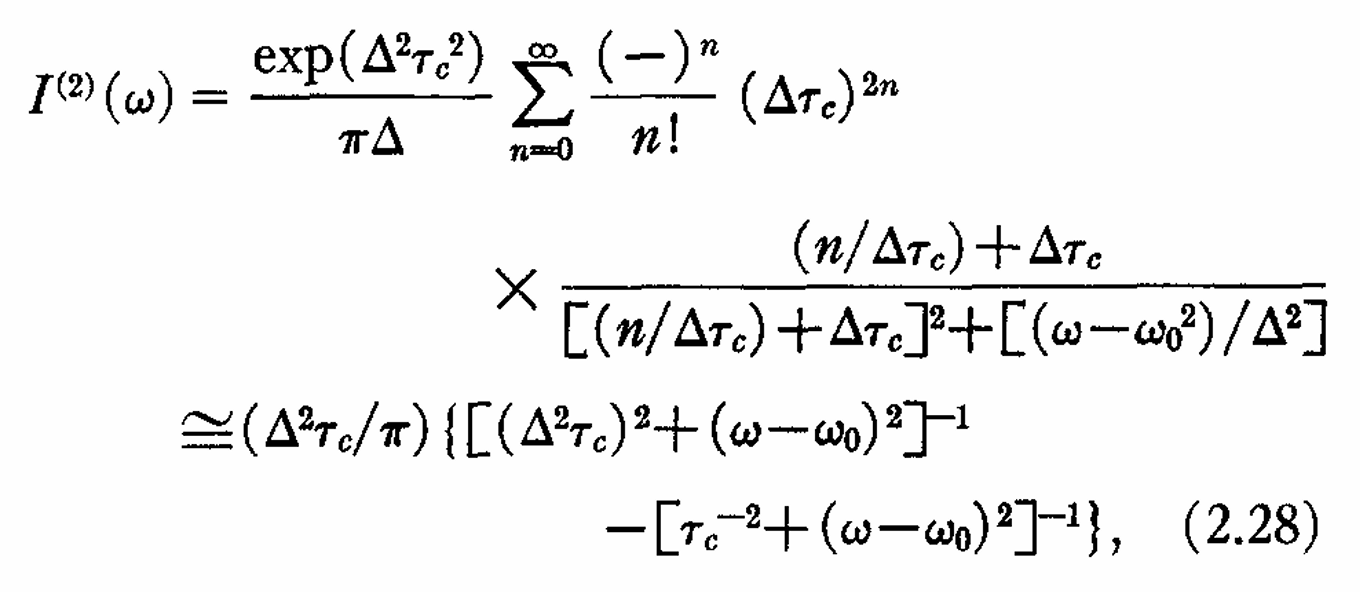
|
|
ABSTRACT: The generalized cumulant expansion method of Kubo is applied to an analysis of spin-relaxation theory appropriate for NMR and ESR studies of molecular systems. It leads to a general, formal solution of the equation of motion of a suitably averaged magnetization operator (and also the spin-density matrix). This solution permits a convenient perturbation expansion in the region of motional narrowing; i.e., when the random functional spin perturbation ℌ1(𝗍) obeys ∣ℌ1(𝗍)∣ τc < 1, where τc is a correlation time for the random process. It is shown how this expansion valid for times 𝗍 ≫ τc generates the time-independent R or relaxation matrix to all orders in ∣ℌ1(𝗍)∣ τc, and detailed expressions are given through fourth order. While the R matrix supplies the linewidth and dynamic frequency shift of the main Lorentzian resonance line, it is found that by formulating the cumulant method without the restriction 𝗍 ≫ τc, weak subsidiary Lorentzian lines are predicted. These subsidiary lines appear as second-order perturbation corrections in ∣ℌ1(𝗍)∣ τc. They have widths given by τc−1 and, in general, large frequency shifts. The problem of (anisotropic)-rotational diffusion is discussed in detail, and it is shown that the spin-Hamiltonian approximation for liquids rests upon a Born–Oppenheimer-type approximation appropriate for random modulation of the nuclear coordinates, i.e., τc−1 ≪ ωn,o, where ℏωn,o is the energy separation between the ground electronic state and the nth excited state coupled by the angular-momentum operator L. The cumulant method is then used to obtain higher-order corrections to the g-tensor line-broadening mechanism.
|
|
|
Electron-Electron Double Resonance of Free Radicals in Solution
J.S. Hyde, J.C.W. Chien, and J.H. Freed.
J. Chem. Phys. 48, 4211-4226 (1968).
<doi: 10.1063/1.1669760>
Publication #20
|
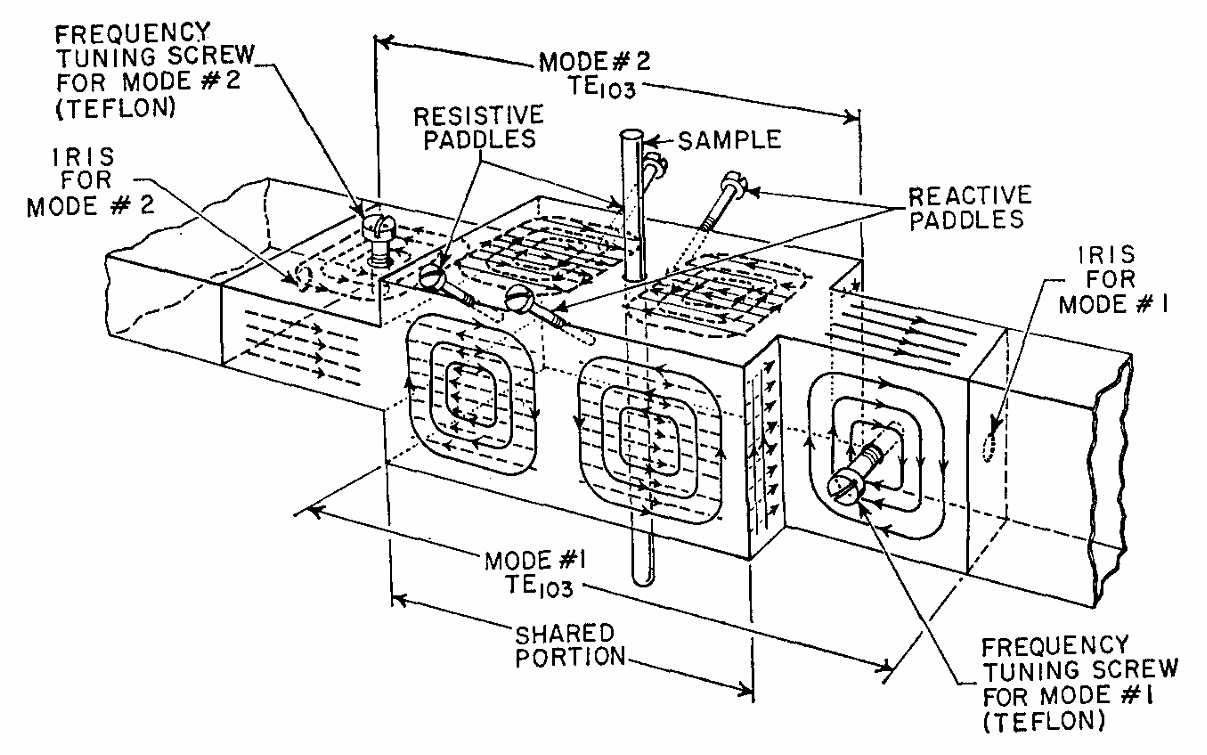
|
|
ABSTRACT: The technique of electron–electron double resonance, in which one part of the EPR spectrum of a paramagnetic sample is irradiated with an intense microwave field and the effect of this intense field on other parts of the spectrum is determined utilizing a second weak microwave field, has been applied to free radicals in solution. EPR signals detected by the weak microwave source are reduced in intensity when the two frequencies are separated by an integral number of hyperfine intervals. In some cases this reduction is as much as 40%. A bimodal cavity of novel design capable of supporting the two resonant microwave modes is described. A nitroxide radical has been investigated in greatest detail. Two mechanisms have been identified: rapid nuclear relaxation induced by electron–nuclear dipolar (END) interaction, which is dominant at low concentrations and temperatures, and Heisenberg exchange (HE), dominant at high concentrations and temperatures. A theoretical analysis of the relaxation processes is presented which permits extracting the critical relaxation parameters from the experimental data. The main theme of the paper is a description of the basic effects, but there would appear to be a number of analytical or structural applications of this technique.
|
|
|
Theory of Saturation and Double Resonance Effects in ESR Spectra. III. Rf Coherence and Line Shapes
J.H. Freed, D.S. Leniart, and J.S. Hyde.
J. Chem. Phys. 47, 2762-2773 (1967).
<doi: 10.1063/1.1712295>
Publication #19
|
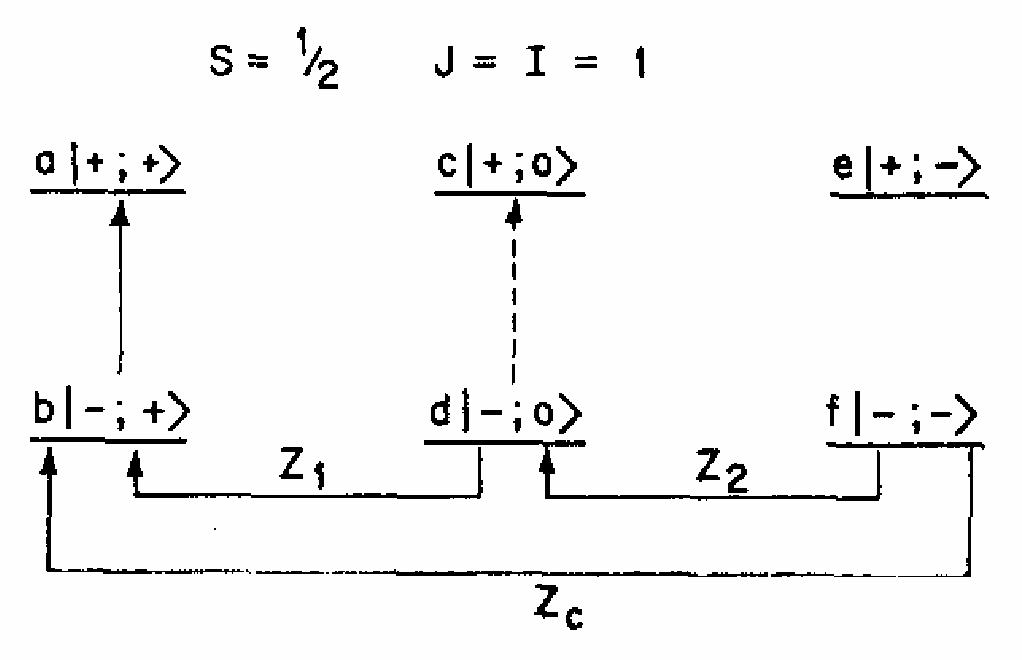
|
|
ABSTRACT: The theory of saturation in the electron spin resonance spectra of dilute solutions of free radicals has been employed to analyze in detail several possible effects in ENDOR spectra that are due to the coherent nature of the applied nuclear rf and microwave fields. These effects are implicitly contained in a "coherence matrix," although their observation also depends on the details of the relaxation terms. It is shown that (1) strong nuclear rf fields can split ESR lines, a phenomenon similar to "spin-tickling" in NMR double resonance, (2) strong microwave fields in the presence of weaker, but saturating, nuclear rf fields can split ENDOR lines, and this latter effect is demonstrated experimentally. A subtle coherence effect requiring nuclear spins of I > ½, or more than one equivalent spin of I = ½, and dependent upon nuclear rf field strength is carefully analyzed. It is predicted to significantly affect only the ENDOR line obtained from saturating the M = 0 hyperfine line for two or four equivalent protons. The theory for this effect appears to be in very good agreement with detailed experimental studies on Coppinger's radical. Other radicals are also found to exhibit such coherence effects. The ability to distinguish and analyze them is deemed important in applying ENDOR as an analytical tool. Computer simulations of the line shapes in the presence of coherence effects are also presented.
|
|
|
Theory of Saturation and Double Resonance Effects in Electron Spin Resonance Spectra. 11. Exchange vs. Dipolar Mechanisms
J.H. Freed.
J. Phys. Chem. 71, 38-51 (1967).
<doi: 10.1021/j100860a006>
Publication #18
|
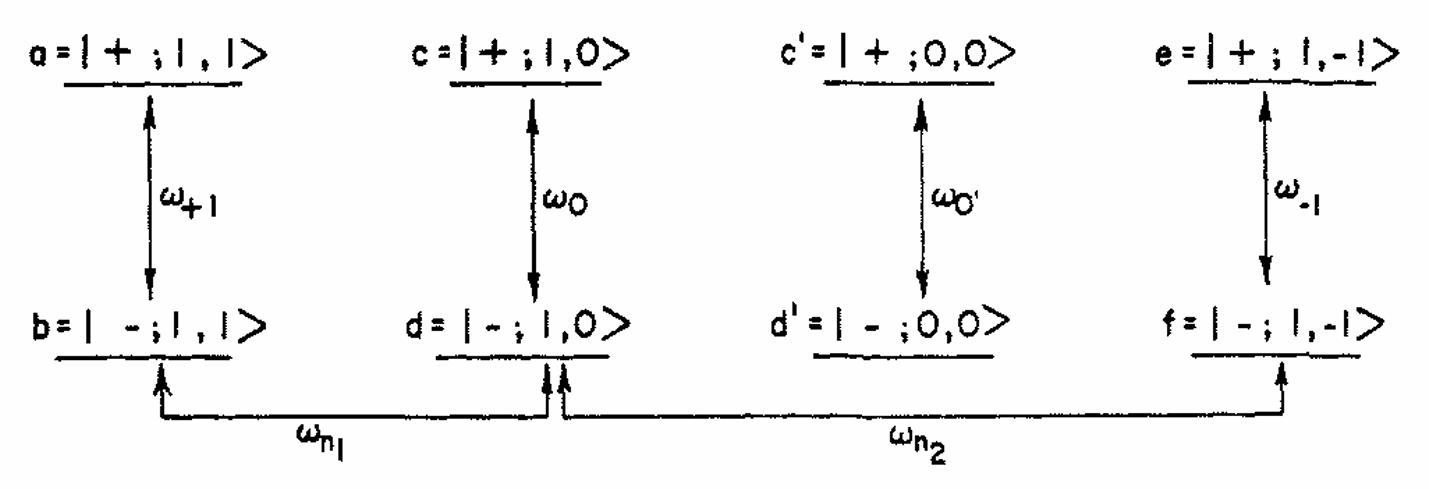
|
|
ABSTRACT: The theory of saturation and line widths in the electron spin resonance spectra of dilute solutions of free radicals has been extended to include incipient effects of chemical exchange and spin-spin or Heisenberg exchange processes as well as the effects of intramolecular terms such as the g-tensor, spin-rotational, and electron-nuclear dipolar (END) interactions. The development is based on an assumption of the statistical independence of the exchange and molecular rotational processes and involves combining the Bloch-Redfield density matrix treatment with the Kaplan–Alexander exchange formalism. It is shown that exchange effects act to average out the differences in widths of components of a composite hyperfine line, so that when the exchange frequency is significantly greater than any such differences, the composite line may be treated as a simple Lorentzian line with an averaged width. In such cases, the esr saturation behavior is again describable in terms of a simple Lorentzian and one may average over the degenerate states. This result is in contrast to the case when exchange effects are weak, while the END effects are important. In an electron-nuclear double resonance (endor) experiment, simple averaging over degenerate states and transitions is no longer adequate because of the specific selection rules of the radiofrequency-induced nmr transitions, and the appropriate modifications are discussed. It is found that a chemical exchange mechanism, in which the diamagnetic species are polarized by the radicals under esr saturation, has effects identical with Heisenberg exchange on any spectra with well-resolved hyperfine structure. They both affect esr saturation parameters as though they yield nuclear-spin transitions between all pairs of states for which ΔMs = 0. However, they do not lead to endor enhancements, but rather are found to diminish the enhancements resulting from other processes. If the diamagnetic species remain unpolarized by the radicals under esr saturation, a situation that does not appear likely, then the exchange would lead to endor enhancements, which, however, could in many cases be distinguished from those induced by an END mechanism. The endor experiments of Hyde are discussed and are found to agree well with the predictions that the END mechanism is the dominant nuclear spin flip process and that exchange effects act to reduce enhancements. Temperature-dependent effects on enhancements are also considered.
|
|
|
On Heisenberg Spin Exchange in Liquids
J.H. Freed.
J. Chem. Phys. 45, 3452-3453 (1966).
<doi: 10.1063/1.1728131>
Publication #17
|
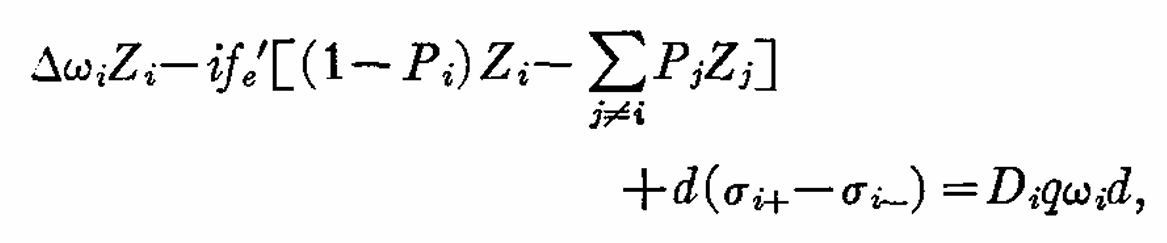
|
|
INTRODUCTION: IT has recently come to our attention that the two theoretical treatments that have been developed for the analysis of the effects of spin-spin or Heisenberg exchange on ESR spectra of free radicals in dilute liquid solutions, must, in fact, be mutually incompatible. Thus, while both theories predict an initial broadening followed by a coalescence of the hyperfine structure into a single exchange-narrowed line, the detailed expressions for the spin exchange are different. The Kivelson treatment of the initial broadening (which bears some similarity to Bloembergen's treatment of the somewhat different problem of a spin-exchange interaction between paramagnetic ions and solvent protons) separates the problem into a secular effect (from the J S1z S2z terms) and a nonsecular effect (from the J S1± S2∓ terms) and finds for example that, when a2 τ12 ≪ 1, where τ1 is a mean encounter time and a the hyperfine splitting, both effects contribute equally to the incipient broadening of the hyperfine lines. One disadvantage of Kivelson's approach is that it cannot describe the region intermediate between good hyperfine resolution and strong narrowing. Currin's treatment can cover the complete range, although the formalism is somewhat unfamiliar and the physical nature of some of the mathematical assumptions is not always clear. Currin examines carefully the case when J, τ1−1 ≪ a and obtains results which suggest that the spin-exchange broadening mechanism is purely a nonsecular effect. That is, following Pake and Tuttle, one may define an effective exchange frequency ƒe = p ∕ τ2, where τ2 is a mean time between collisions and p is the probability of exchange per encounter of the radical pairs. But the p as obtained by Currin is just that which may be calculated simply from the well-known quantum-mechanical solution to transitions induced under the action of a constant perturbation taken as J S1± S2∓. Thus the spin exchange becomes a lifetime-broadening-only mechanism in contrast to more typical linewidth phenomena which have important secular (or frequency-modulation) effects. This was a conclusion reached earlier by Wittke and Dicke in their treatment of gas-phase spin exchange of H atoms in low fields.
|
|
|
Theory of Spin Relaxation Via Quantum-Molecular Systems: Resonance Effects
J.H. Freed.
J. Chem. Phys. 45, 1251-1257 (1966).
<doi: 10.1063/1.1727744>
Publication #16
|
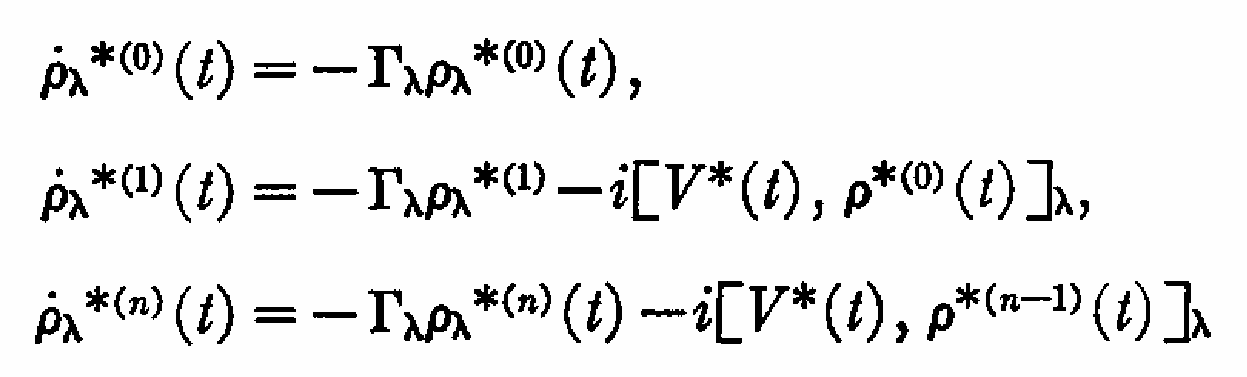
|
|
ABSTRACT: A relaxation equation is derived for the spin-density matrix on the assumptions that (1) the spins are weakly coupled to the molecular degrees of freedom which are described quantum mechanically and (2) the relaxation of the molecular systems is well represented by a general time-independent relaxation matrix which can include effects of weak as well as strong collisions. The irreversibility of the spin relaxation is achieved with no further assumptions, and is given in detail in terms of the normal modes of relaxation of the molecular systems. It is also pointed out that the present formulation inherently includes the assumption of resonant-type interactions between molecular systems and spins.
|
|
|
Theory of Saturation and Double-Resonance Effects in ESR Spectra
J.H. Freed.
J. Chem. Phys. 43, 2312-2332 (1965).
<doi: 10.1063/1.1697128>
Publication #15
|
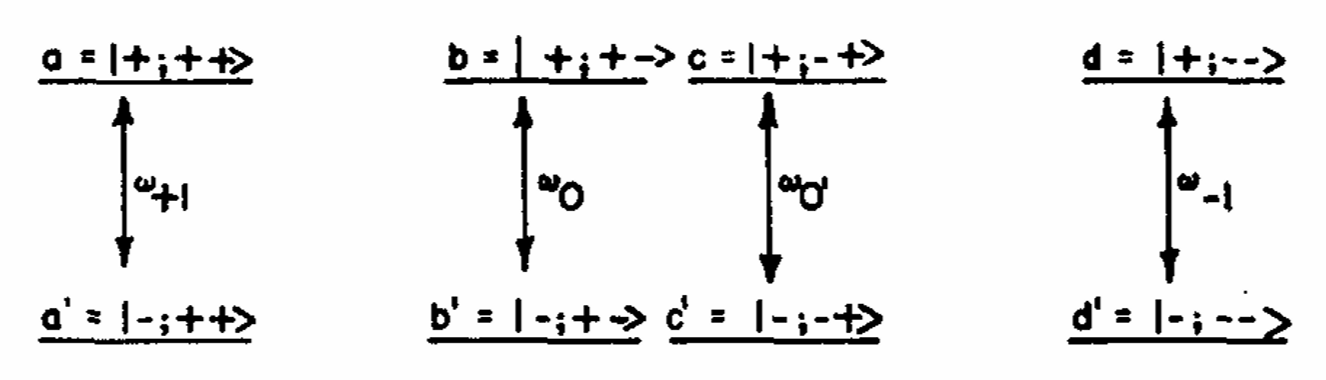
|
|
ABSTRACT: A theory of saturation in the electron spin resonance spectra of dilute solutions of free radicals has been developed in terms of the general Boltzmann equation for the density matrix given by Bloch and Redfield. It is shown, in contrast to the earlier theories, that, in general, a composite line arising from a set of degenerate nuclear spin states must be described by a generalized (matrix) saturated Lorentzian rather than a single line of over-all saturated Lorentzian shape. The general properties of this solution are discussed in detail for the case when electron—nuclear dipolar interactions are the only important nuclear-spin-dependent relaxation mechanism. A particularly simple situation exists when similar nuclei are completely equivalent. Then each composite line consists of a superposition of saturated Lorentzians, and the linewidths and saturation parameters of each component depend only on the values of the total spin and of the z component of spin of the associated configuration of the completely equivalent nuclei. Thus it would be possible to saturate some components to a greater extent than others. These simple components may become coupled together in a complex fashion when the time-dependent fluctuations of the dipolar interactions for equivalent nuclei are no longer identical, which is often the case for aromatic-ring protons. Further coupling may be expected from the effects of nuclear-quadrupole interactions and from intermolecular-exchange phenomena and are only qualitatively discussed. In the absence of saturation the present theory reduces to the linewidth theory of Freed and Fraenkel.
Detailed expressions are obtained for steady-state electron—nuclear double-resonance (ENDOR) effects on radicals containing completely equivalent sets of nuclei. The saturating fields can lead to coherence and induced-transition effects. It is shown how the latter, under appropriate conditions, can result in enhancement of saturated ESR spectra. It appears necessary for enhancements that lattice-induced nuclear spin transitions should be comparable in magnitude to the lattice-induced electron spin transitions, or else cross transitions involving both nuclear and electron spins must be important in the relaxation process. Coherence effects involving both electron and nuclear spin transitions should be unimportant when the main contribution to the ESR linewidths results from secular processes and is nuclear spin independent. This is also the condition for NMR linewidths to be significantly narrower than ESR linewidths. Complications still arise from the coherence and linewidth coupling effects of the different nuclear transitions being excited. A brief discussion of enhancement by a transient "heating" effect is also given.
|
|
|
Quantum Effects of Methyl-Group Rotations in Magnetic Resonance: ESR Splittings and Linewidths
J.H. Freed.
J. Chem. Phys. 43, 1710-1720 (1965).
<doi: 10.1063/1.1696995>
Publication #14
|
|
|
ABSTRACT: The effects of internal rotations of methyl groups on ESR hyperfine lines are analyzed in terms of a quantum-mechanical description of the motion. The classical description of rotational averaging is replaced by (1) spin—rotational coupling from a hyperfine operator, and (2) rotational relaxation by thermal collisions. In the absence of collisions, the effects of (1) lead to a static set of splittings whose values depend upon the relative magnitudes of the hyperfine-versus-tunneling frequencies. In the presence of frequent collisions, represented by a strong-collision model, the effects of (1) lead to line broadening, the detailed nature of which also depends on the hyperfine-versus-tunneling frequencies. In general, results predicted from a classical treatment of the motion are obtained when the hyperfine frequency is significantly greater, while quantum effects become important for relatively larger tunneling frequencies. The results are illustrated by application to relevant experimental observations. In the Appendix, the strong-collision relaxation theory is generalized to apply to the present case where the spin—rotational coupling connects states of different nuclear spin symmetry.
|
|
|
ESR Hyperfine Linewidths from Benzene Anion Distortions
J.H. Freed.
J. Chem. Phys. 43, 1427-1428 (1965).
<doi: 10.1063/1.1696941>
Publication #13
|
|
|
ABSTRACT: It was reported several years ago by Townsend and Weissman that the proton hyperfine structure in the paramagnetic resonance of the anions of benzene, triphenylene, and coronene is unusually broad when compared to the sharp hyperfine structure obtained from most other aromatic hydrocarbons studied under identical conditions. They suggested this broadening may be associated with dynamic Jahn-Teller distortions which would modulate the spin distributions. McConnell and McLachlan (MM) presented a detailed calculation on the benzene negative ion to demonstrate that its ground vibronic state is doubly degenerate, and they attributed the anomalously broad benzene anion linewidths to the effects of a fluctuating Stark effect from the polar solvent. They argue that this solvent interaction will remove the ground-state vibronic degeneracy so that the lower energy state is approximated by the molecular orbital
ψ = ½ (Φ1 + Φ2 − Φ4 − Φ5). (1)
(Their comments on the spectrum, as well as those presented here are not radically altered by the other choice.) Equation (1) predicts one-quarter of an unpaired electron on each of four carbon atoms and zero on the opposite carbons, 3 and 6. However, the solvent effect, which is random, would shift the distortion so that at different times the nodes are at Carbons 2 and 5 and also at 1 and 4. MM point out that when the switching time τ obeys the condition τ ≪ 1∕2πΔν, where Δν is the hyperfine splitting (|Q| ∕ 4 = 15.7 Mc/sec)3for one of the distorted configurations, that a single spectrum averaged over the three configurations is obtained, and this is consistent with the observed hyperfine splittings of |Q| ∕ 6 and the intensity distribution of 1:6:15:20:15:6:1. According to MM the broad lines can still arise from the residual effect of the rapid switching, resulting in a line broadening, Δνh ˜(Q ∕ 6)22πτ, so τ ˜10−9 sec.
|
|
|
Semiclassical Theory of the Effects of Internal Motions on the Linewidths in Electron Spin Resonance Spectra
J.H. Freed and G.K. Fraenkel.
J. Chem. Phys. 41, 3623-3638 (1964).
<doi: 10.1063/1.1725777>
Publication #12
|
|
|
ABSTRACT: A number of classical dynamical models are developed for describing the effects of internal rotational motions and solvent-complex formation on the ESR hyperfine linewidths of free radicals in solution. These dynamical processes can lead to linewidth effects because they cause time-dependent modulations of the isotropic hyperfine interactions of the different magnetic nuclei. An alternating linewidth effect, which has been observed in a number of recent studies, is predicted to result when there is an out-of-phase correlation between the hyperfine splittings ai(t) of equivalent nuclei. By equivalent nuclei here are meant those for which the time-average splittings ‹ai(t)›Av are equal, and an out-of-phase correlation is one in which an increase in the instantaneous splitting from Nucleus i, ai(t), is correlated with a decrease in the splitting aj(t) from nucleus j. This out-of-phase correlation can result from a coupling of the mechanical motions of different rotating groups and also from the effects that changes in orientations of uncorrelated rotors may have on redistributing electron spin density in the molecule. Continuous motion cases are treated by Brownian-motion theory using coupled and uncoupled internal rotors and torsional oscillators. Random jump models between discrete states are also considered. It is shown that all these models can give rise to an alternating linewidth effect provided that certain definite relationships of the proper type exist for the coupling of the motions or the variations of the spin densities. For many of these models the correlation a1(t) = a2(t) for all t, or complete equivalence of both nuclei, may also result when different relationships exist. The correlation of the splittings of more than two groups of completely equivalent nuclei will, in general, lead to more complex linewidth effects.
|
|
|
Anisotropic Rotational Diffusion and Electron Spin Resonance Linewidths
J.H. Freed.
J. Chem. Phys. 41, 2077-2083 (1964).
<doi: 10.1063/1.1726208>
Publication #11
|
|
|
ABSTRACT: The recent general theory of ESR linewidths for dilute solutions of free radicals developed by Freed and Fraenkel is extended to cover the possibility that the rotational motions of the free radicals are anisotropic. The analysis is greatly simplified by expressing the solution of the rotational-diffusion equation as an expansion in Wigner rotation matrices. Only the spectral-density functions in the linewidth expressions given by Freed and Fraenkel are changed by the inclusion of anisotropic rotational diffusion. These spectral-density functions are found to depend upon five different relaxation times which are expressible in terms of the three principal values of the diffusion tensor ℜ. It is shown how an analysis of the ESR linewidth effects, which vary from one hyperfine component to another, may be used in conjunction with calculations of the anisotropic electron—nuclear dipolar interactions to estimate the principal values of ℜ. The experimental linewidth study on the para-dinitrobenzene anion radical is reanalyzed, and it is shown that an anisotropic rotational-diffusion mechanism cannot by itself explain the anomalous absence of any significant quadratic linewidth dependence on the total Z component of spin of the ring protons. While an order of magnitude estimate of ℜ was made, further information permitting a quantitative understanding of the anomalous result (presumably due to fluctuations in isotropic hyperfine interactions) would be necessary to obtain a meaningful estimate of the principal values of ℜ for this radical. The physical significance of ℜ in the approximation of a Brownian particle as well as in terms of intermolecular potentials is discussed.
|
|
|
Theory of Line Widths in Electron Spin Resonance Spectra: Motion of Methyl Groups
J.H. Freed and G.K. Fraenkel.
J. Am. Chem. Soc. 86, 3477-3484 (1964).
<doi: 10.1021/ja01071a016>
Publication #10
|
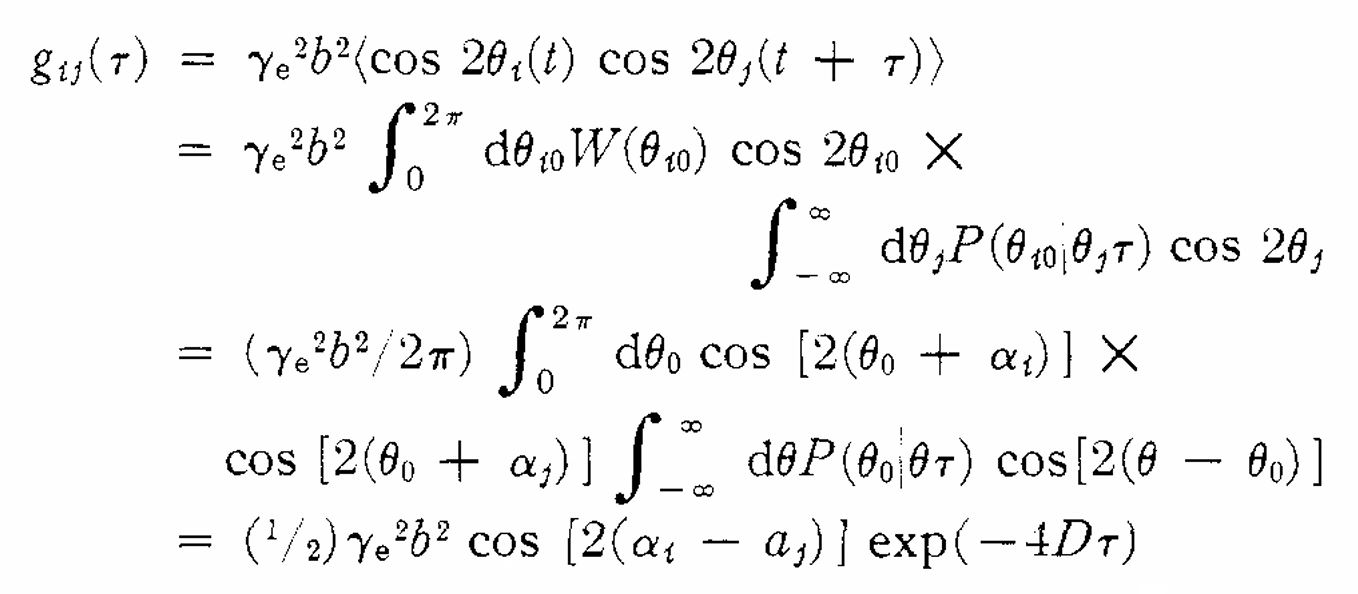
|
|
ABSTRACT: The relaxation-matrix theory of line widths in electron spin resonance spectra has been employed to analyze the line-width effects arising from the motions of methyl groups in π-electron free radicals. Broadening of the lines can result because the methyl-proton hyperfine splittings are a function of the angle of orientation of the methyl group, and thus the splittings fluctuate as this angle varies. The motion of the methyl groups is treated by a Brownian motion model assuming a free rotatory diffusion about the C–C bond between the methyl group and the aromatic system to which it is bonded, and also by a jump model in which the group can undergo transitions from one to another of three different equilibrium orientations. Radicals with several methyl groups are analyzed as having either completely correlated or completely uncorrelated motions. The correlated motions are approximated by assuming a gear-like interleaving without slip of the hydrogen atoms on methyl groups substituted at adjacent positions on an aromatic ring. When the motions of the methyl group cause large contributions to the line width, the central pair of lines from the splittings by one methyl group are predicted to be broad and the end lines narrow. When there are two or four methyl groups in the radical, large linewidth contributions lead to a spectrum in which every third line is narrow while the remaining lines are broad, an effect which is analogous to the alternating line-width phenomenon. For very rapid rotations, nonsecular as well as secular line-width contributions are important, and consequently the nondiagonal relaxation matrix for the case of one methyl group has been analyzed in detail. None of these effects of methyl-group rotations has been observed in the e.s.r. spectra of aromatic radicals, and from the negative results it is possible to estimate that the relaxation time τc for the rotation of the methyl groups is less than 10−8 sec. The predicted effects are most likely to be found by low-temperature studies on radicals which have large methyl-proton hyperfine splittings and highly hindered methyl groups.
|
|
|
Line Shapes in Electron Spin Resonance Spectra
J. Gendell, J.H. Freed, and G.K. Fraenkel.
J. Chem. Phys. 41, 949-959 (1964).
<doi: 10.1063/1.1726038>
Publication #9
|
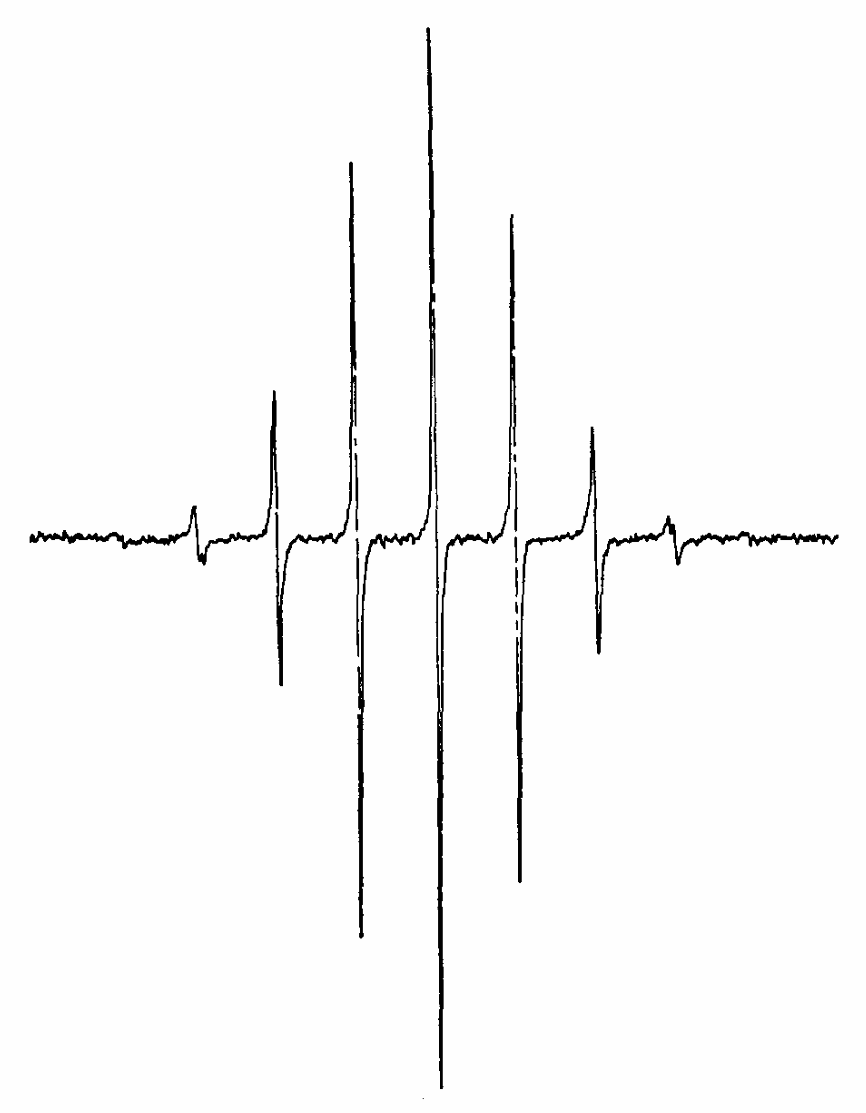
|
|
ABSTRACT: The theory of the linewidths in the electron spin resonance spectra of free radicals recently developed by Freed and Fraenkel predicts that, in general, composite lines arising from a set of degenerate nuclear spin states should not be Lorentzian in shape, and that the shapes of different lines in the same spectrum should be different. The earlier theory of Kivelson predicted that all the lines should be Lorentzian. To test the differences between the two theories, experimental studies of the line shape in the spectrum of the tetracyanoethylene anion have been made in a solvent consisting of a mixture of absolute ethanol and glycerine. Different lines were found to have different shapes, and the shapes were well represented by a sum of Lorentzian-shaped components, in agreement with the newer theory. Studies of dimethylsulfoxide solutions of the p-benzosemiquinone ion were also made. The spectra obtained in this system show only small linewidth variations and, in agreement with the theory, the variations in line shape are negligible. It was also possible to demonstrate that the nitrogen hyperfine splitting aN in the tetracyanoethylene radical is positive.
|
|
|
Alternating Linewidths and Related Phenomena in the Electron Spin Resonance Spectra of Nitro-Substituted Benzene Anions
J.H. Freed and G.K. Fraenkel.
J. Chem. Phys. 41, 699-716 (1964).
<doi: 10.1063/1.1725949>
Publication #8
|
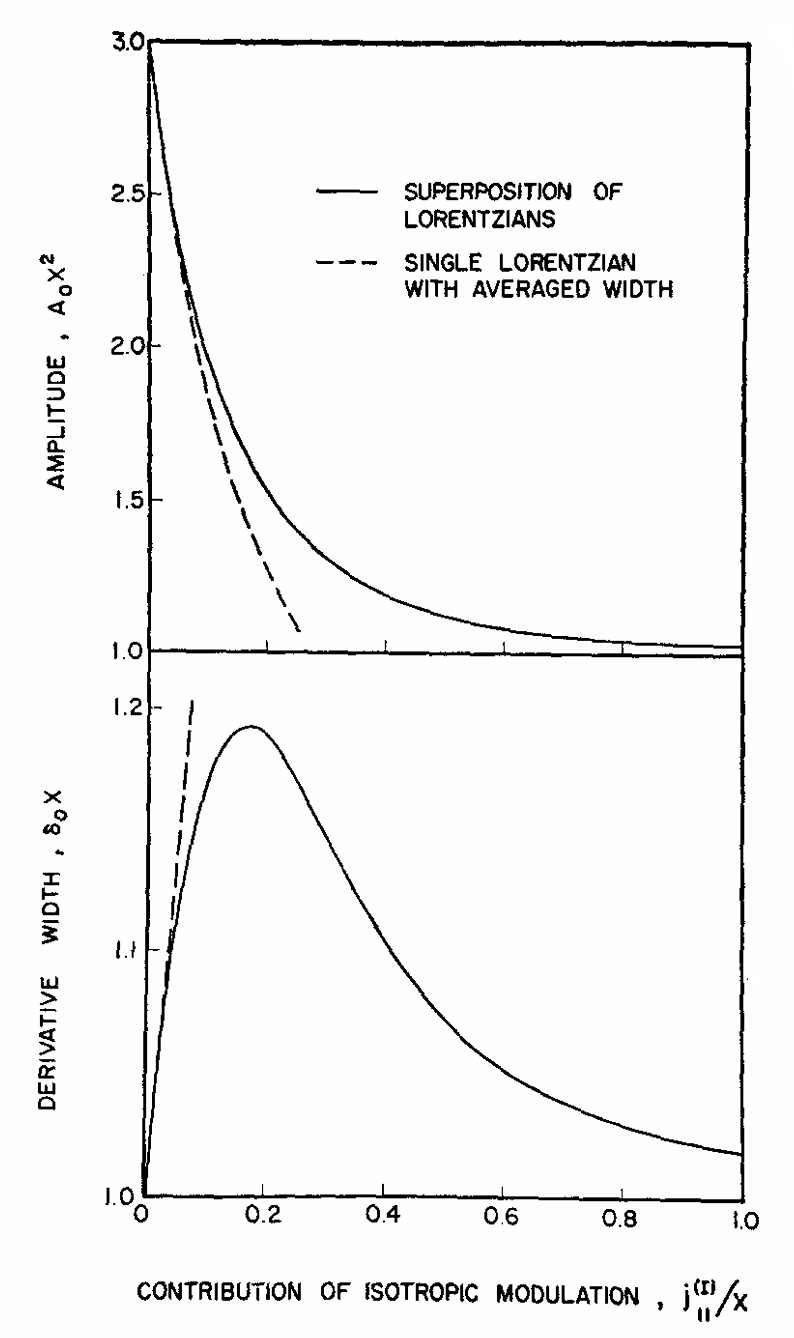
|
|
ABSTRACT: Studies of linewidths in the electron spin resonance spectra of a number of nitro-substituted benzene anions are reported. The radicals were generated electrolytically in N,N-dimethylformamide solutions, and spectra were obtained over a range from above to below room temperature. Pronounced alternations in the linewidths of the nitrogen lines in many of the spectra were observed. If MN is the total z component of the nuclear spin angular momentum in compounds containing two equivalent nitrogen nuclei, the alternating linewidth phenomenon causes a broadening of the lines for which MN = ± 1 and a reduction in the amplitude of the central line. Large alternating linewidths were observed in the room-temperature spectra of the anions of dinitrodurene, dinitromesitylene, and 2,6-dinitro-3,5-dimethyl-4-acetyl-t-butylbenzene; and also in a spectrum which shows splittings from two equivalent nitrogen nuclei that was obtained from trinitromesitylene. Slight alternating linewidth effects at room temperature that became more marked at low temperatures were found in the spectra of the 2,6-dinitrotoluene and m-dinitrobenzene anions. No linewidth alternations were detectable even at low temperatures in the spectra of the o-dinitrobenzene, p-dinitrobenzene, and 2,6-dinitrophenolate anions. The 2,6-dinitroaniline anion, which was investigated only at room temperature, showed no evidence of an alternating linewidth effect. None of the spectra of the mononitrobenzene anions examined exhibited any anomalous linewidth phenomena. The experimental observations are in general agreement with the recently developed theory of linewidths. The theory attributes the alternation in widths in these radicals to an out-of-phase correlation of a modulation of the isotropic hyperfine splittings of the two equivalent nitrogen nuclei. This out-of-phase modulation probably arises from either fluctuating solvent complexes with the nitro groups, or from internal rotations of these groups relative to the plane of the benzene ring, and both types of motion may occur simultaneously. Steric hindrance of the nitro groups was found to enhance the magnitude of the alternations in width, as did lowering of the temperature.
|
|
|
On the Theory of Spin Relaxation of Gas Molecules: The Strong-Collision Limit
J.H. Freed.
J. Chem. Phys. 41, 7-13 (1964).
<doi: 10.1063/1.1725651>
Publication #7
|
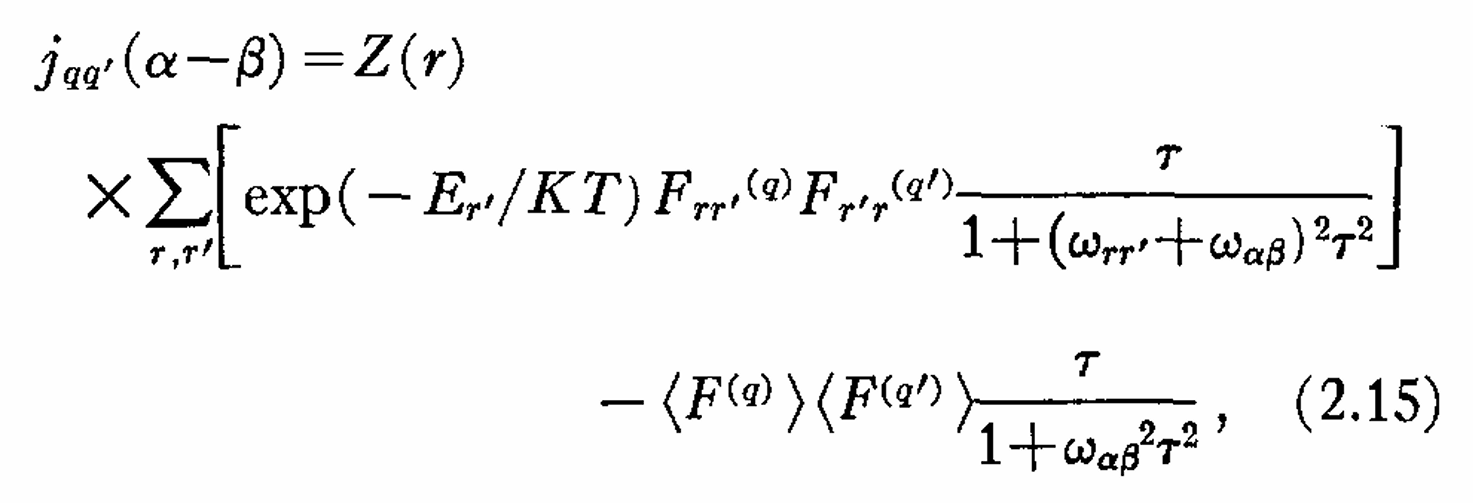
|
|
ABSTRACT: A model of spin relaxation of gas molecules based upon an assumption of strong collisions is developed. A relaxation matrix formally similar to that of Redfield, but with different spectral densities, is obtained. The nuclear magnetic relaxation of gaseous orthohydrogen is discussed in terms of these results, and it is shown that, in general, molecules in different J states are expected to exhibit different relaxation times (a conclusion that is independent of the strong collision assumption) whenever the collision mechanism is effective only in reorienting J but not in changing its magnitude. Otherwise statistical averages of relaxation times are obtained. Relaxation of more complex molecules is also discussed.
|
|
|
Linewidth Studies in Electron Spin Resonance Spectra: The Para and Ortho Dinitrobenzene Anions
J.H. Freed and G.K. Fraenkel.
J. Chem. Phys. 40, 1815-1829 (1964).
<doi: 10.1063/1.1725411>
Publication #6
|
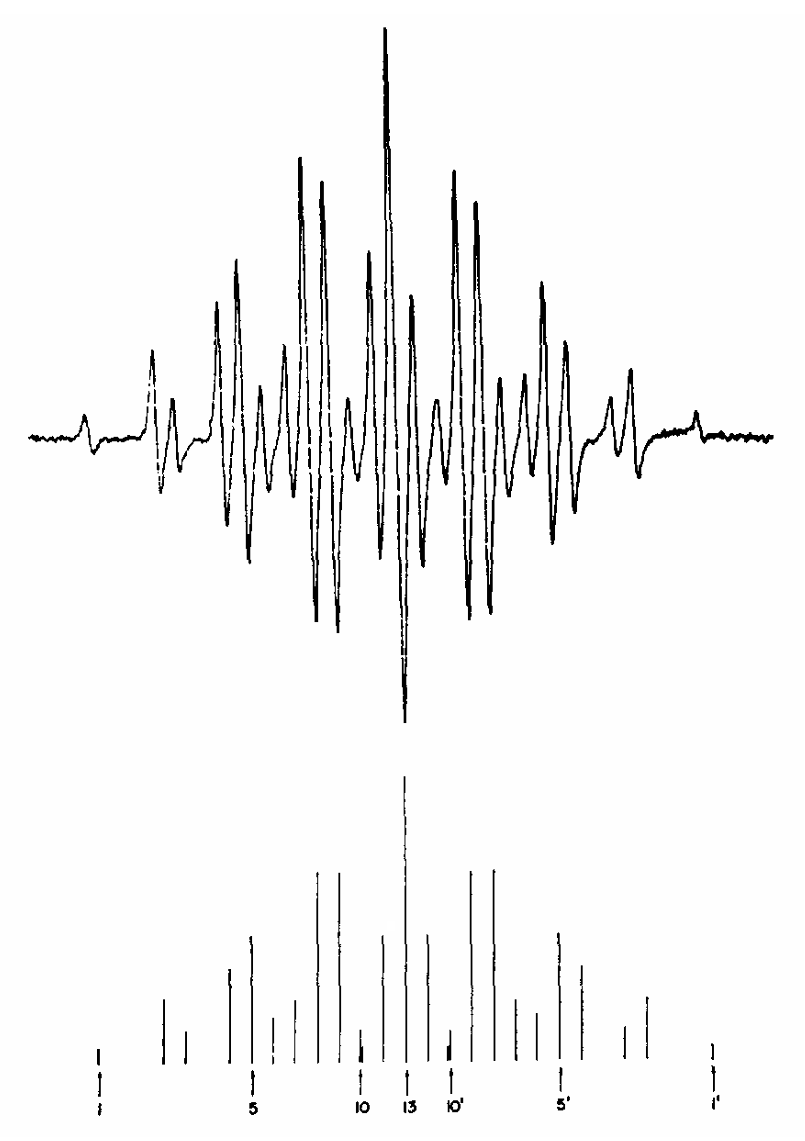
|
|
ABSTRACT: Large variations have been found among the linewidths of the different hyperfine lines in the low-temperature electron spin resonance spectra of the p- and o-dinitrobenzene anions generated electrolytically in N,N-dimethylformamide solutions. The magnitude of the variations in the para compound is so great that the spectrum is superficially uninterpretable. Detailed analysis of the spectrum of this radical shows, however, that most of the linewidth differences can be accounted for by modulation through molecular tumbling of the intramolecular anisotropic dipolar and g-tensor interactions. There may also be a small contribution from modulation of the isotropic proton hyperfine splittings, the mechanism that accounts for the alteranting linewidth phenomenon in a number of radicals, but an alternation of the widths is not observed in the p-dinitrobenzene anion spectrum because the contribution from this interaction is small. The sign of the isotropic nitrogen hyperfine splitting aN has been determined by a new method involving only pure dipolar interactions with the nitrogen nuclei and protons and not depending on any assumptions about the magnitudes of the components of the g tensor. A number of features of the relaxation matrix determining the linewidths are discussed, and the problems encountered when the components of a degenerate line have different widths are analyzed. The spectra obtained in dimethylformamide solutions are much better resolved than those previously reported in acetonitrile solutions. Changes of hyperfine splittings with both solvent and temperature are observed.
|
|
|
Theory of Linewidths in Electron Spin Resonance Spectra
J.H. Freed and G.K. Fraenkel.
J. Chem. Phys. 39, 326-348 (1963).
<doi: 10.1063/1.1734250>
Publication #5
|
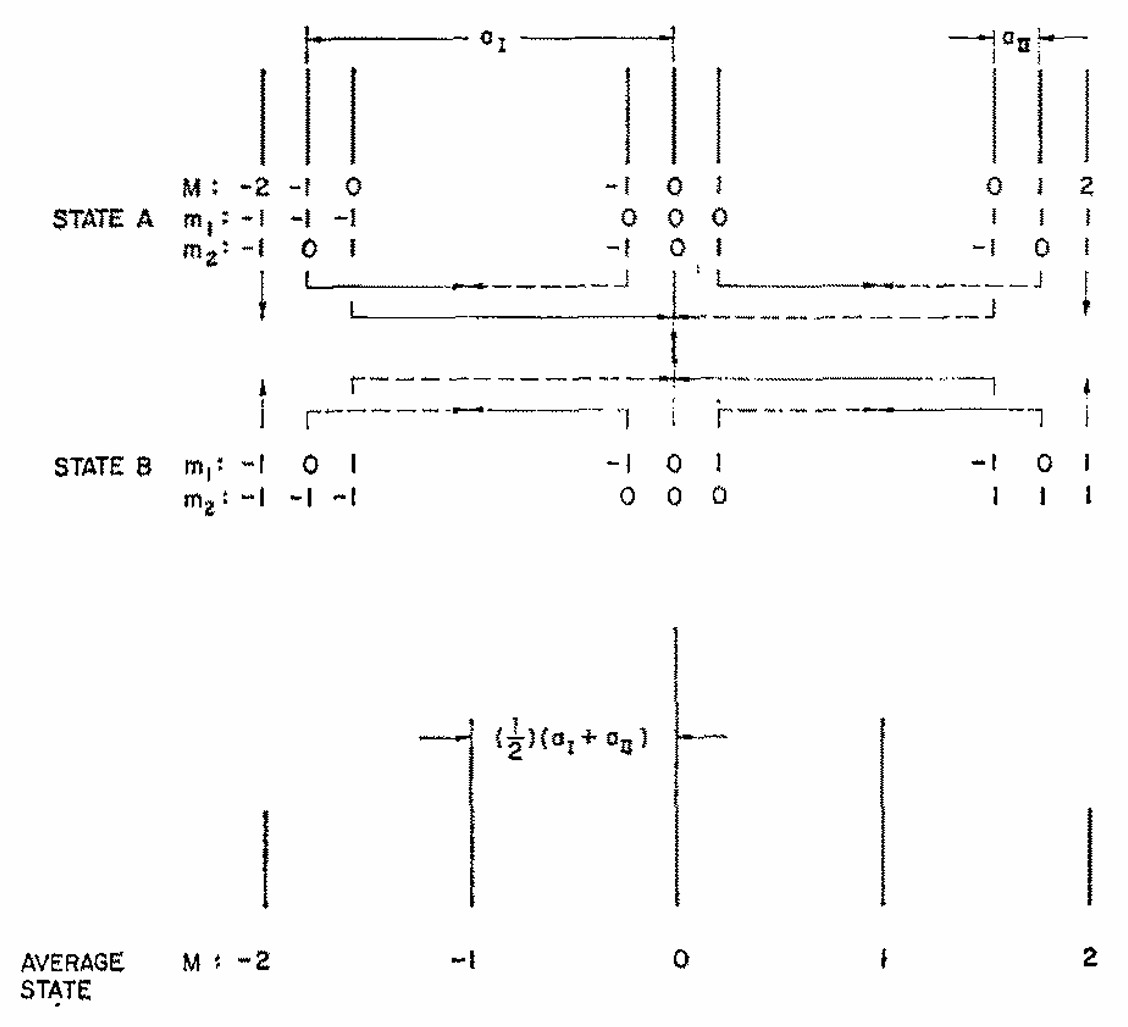
|
|
ABSTRACT: A general theory of the linewidths in the electron spin resonance spectra of dilute solutions of free radicals has been developed in terms of the relaxation-matrix theory of Bloch, Redfield, and Ayant. In contrast to previous theories, it is shown that a composite line arising from a set of degenerate nuclear-spin states should, in general, consist of a sum of superimposed lines of Lorentzian shape with different widths rather than a single line with an over-all Lorentzian shape. A single Lorentzian line is still obtained, however, as a limiting case when the variation of the widths of the different components of a composite line is small compared to the average width. Although the non-Lorentzian shape of a composite line is often difficult to observe experimentally, a number of other observable properties are predicted by the present development that are outside the scope of the previous theories. For example, linewidth effects resulting from differences in the widths of the separate components of a composite line are predicted that explain the alternation in the linewidths from one hyperfine line to another recently observed in the ESR spectra in certain free radicals. The detailed form of the relaxation matrix is presented for intramolecular anisotropic and isotropic electron—nuclear dipolar interactions, quadrupole interactions, and g-tensor relaxations. Modulations of the spin density and hyperfine splittings are included, as are internal motions, and a number of cross terms between the different relaxation mechanisms arise. In general the relaxation matrix of a composite line contains significant off-diagonal elements, and the determination of the linewidths requires the evaluation of the eigenvalues of the matrix. Problems involving rapid chemical exchange, or modulation by jumps to a small number of sites, can be treated by the relaxation-matrix theory and, under special restrictions, by either the modified Bloch equations or the Anderson theory of motional narrowing. When applicable, these latter procedures can be used over the entire range of exchange rates, while the relaxation-matrix theory is limited to fast rates only.
|
|
|
Intramolecular Hydrogen Bonding in ESR Spectra
J.H. Freed and G.K. Fraenkel.
J. Chem. Phys. 38, 2040-2041 (1963).
<doi: 10.1063/1.1733927>
Publication #4
|
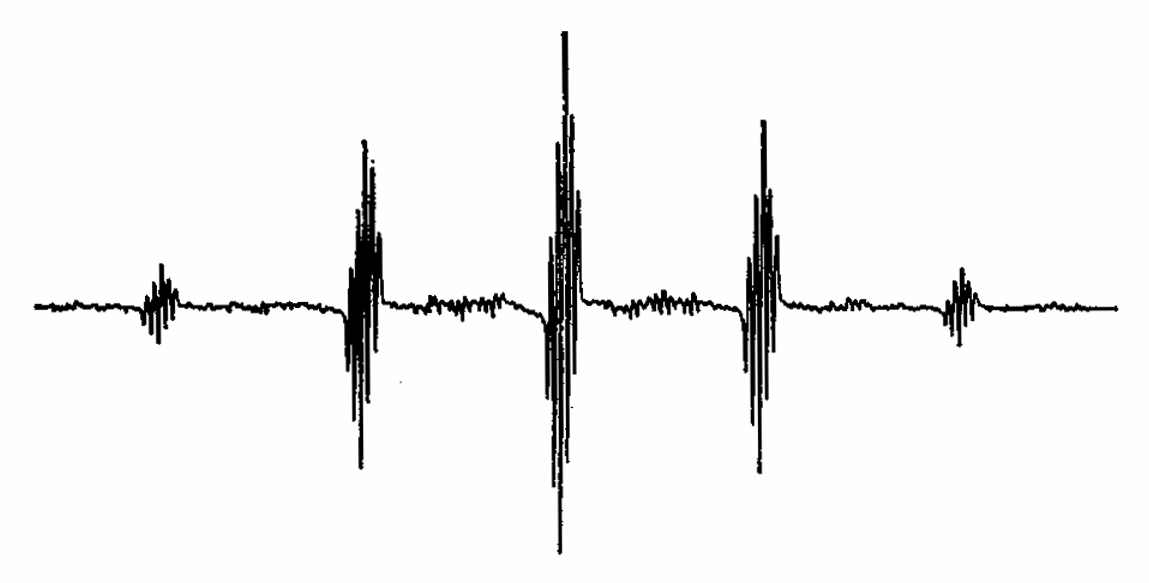
|
|
ABSTRACT: It is well known that naphthazarin (1,4-dihydroxy-5,8-naphthoquinone) and quinizarin (1,4-dihydroxy-9, 10-anthraquinone) exhibit intramolecular hydrogen bonding. We have performed electron spin resonance studies which show that the anion radicals of these compounds also contain hydrogen bonds and that the splitting constants have several interesting and unexpected characteristics.
|
|
|
Solvent Effects in Electron Spin Resonance Spectra
J. Gendell, J.H. Freed, G.K. Fraenkel.
J. Chem. Phys. 37, 2832-2841 (1962).
<doi: 10.1063/1.1733110>
Publication #3
|
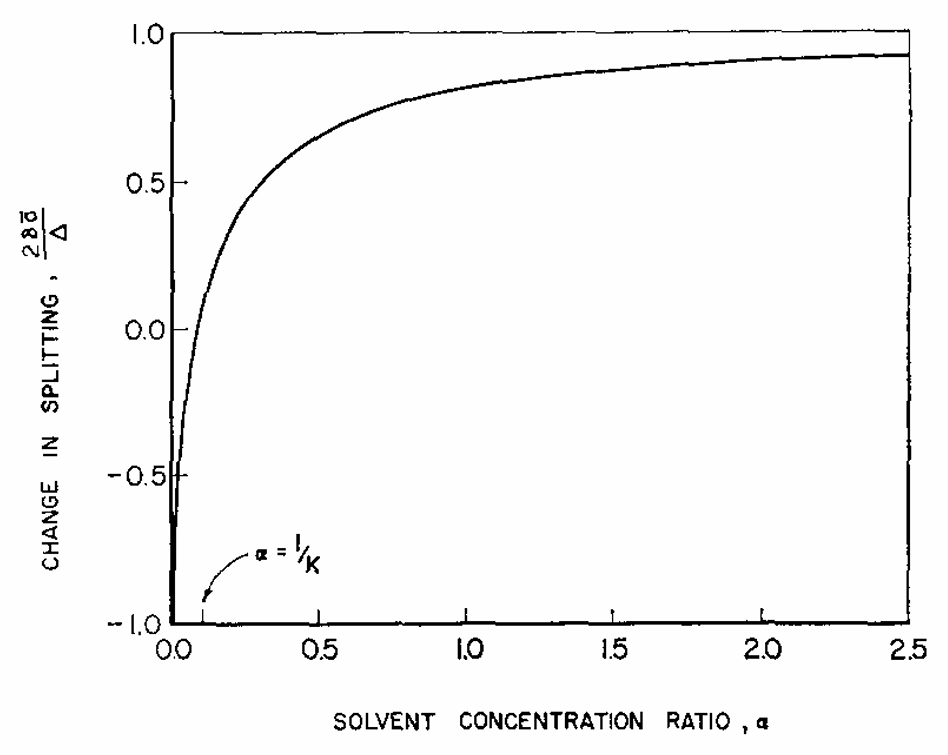
|
|
ABSTRACT: It has been shown that a simple model can account for the available data on the solvent dependence of the hyperfine splittings in the ESR spectra of organic free radicals. We have assumed that the changes in splittings arise entirely from a redistribution of the pi-electron spin density, and that the spin density is affected only by localized complexes between the solvent and polar substituents or heteroatoms in the radical. This model predicts that the magnitudes of the changes in proton splittings should often be small, although large fractional changes at positions of small spin density can sometimes occur. The large variations found for the nuclei of many electron atoms are shown to arise because their splittings are very critical functions of the spin density.
The effect of the solvent on the proton hyperfine splittings in the semiquinones has been treated by assuming that the solvent alters the electronegativity of the oxygen atoms and by performing molecular-orbital calculations to estimate spin density changes. These calculations reproduce both the directions and magnitudes of the changes in proton splittings with solvent and are in agreement with the very large fractional changes observed at some positions with small splittings.
An analysis of the effects on the ESR spectra of the exchange reactions between the different solvent complexes has shown that the spectra normally observed result from systems undergoing rapid exchange. A simple model for a radical with only one functional group which can interact with the solvent (e.g., the nitrobenzene anion), is shown to account very well for the observed variations in hyperfine splittings as a function of the composition of a binary solvent mixture. The treatment of the exchange reactions by use of the modified Bloch equations is compared with the spectral density method. The contribution to the linewidth from solvent-induced fluctuations in the spin-density distribution is calculated for a simple two-jump case, and other factors affecting the linewidths of radicals subjected to random solvent interactions are discussed.
|
|
|
Alternating Linewidths in the ESR Spectra of Dinitrobenzene Anion Radicals
J.H. Freed, P.H. Rieger, and G.K. Fraenkel.
J. Chem. Phys. 37, 1881-1882 (1962).
<doi: 10.1063/1.1733384>
Publication #2
|
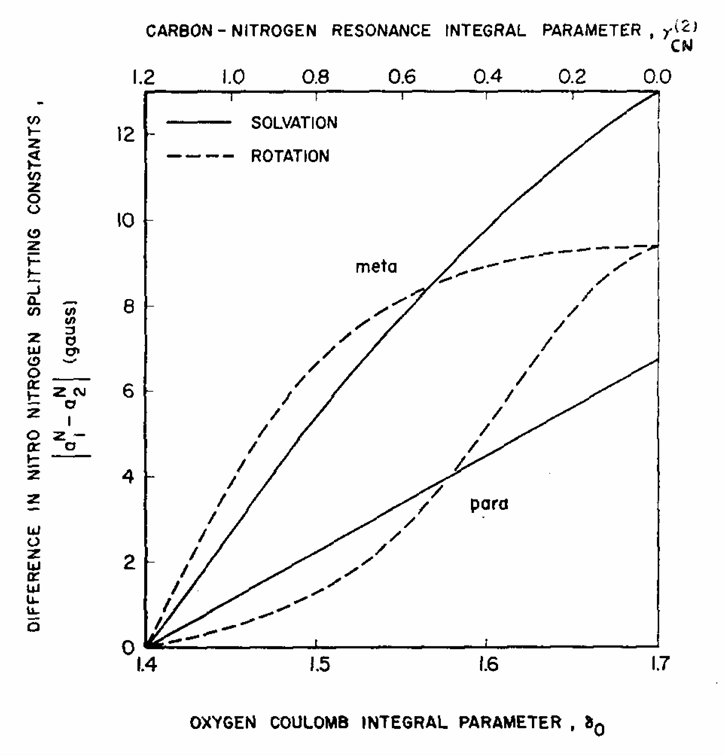
|
|
ABSTRACT: We have found an alternation in the linewidths in the ESR spectrum of the anion radical of m-dinitrobenzene. The radical was produced by electrolytic reduction in N,N-dimethyl-formamide solution at room temperature. The derivative spectrum obtained at −50°C shows that those hyperfine components corresponding to mN = ± 1 (where mN is the total z component of the nitrogen nuclear spin angular momentum) are considerably broader and of smaller amplitude than those for which mN = ± 2.0. At room temperature, the effect is much weaker but may be detected by careful analysis of the spectrum. Examination of the spectra of the p- and o-dinitrobenzene anions under identical conditions failed to show any indication of alternating linewidths.
|
|
|
Anomalous Alternating Linewidths in ESR Spectra
J.H. Freed and G.K. Fraenkel.
J. Chem. Phys. 37, 1156-1157 (1962).
<doi: 10.1063/1.1733238>
Publication #1
|
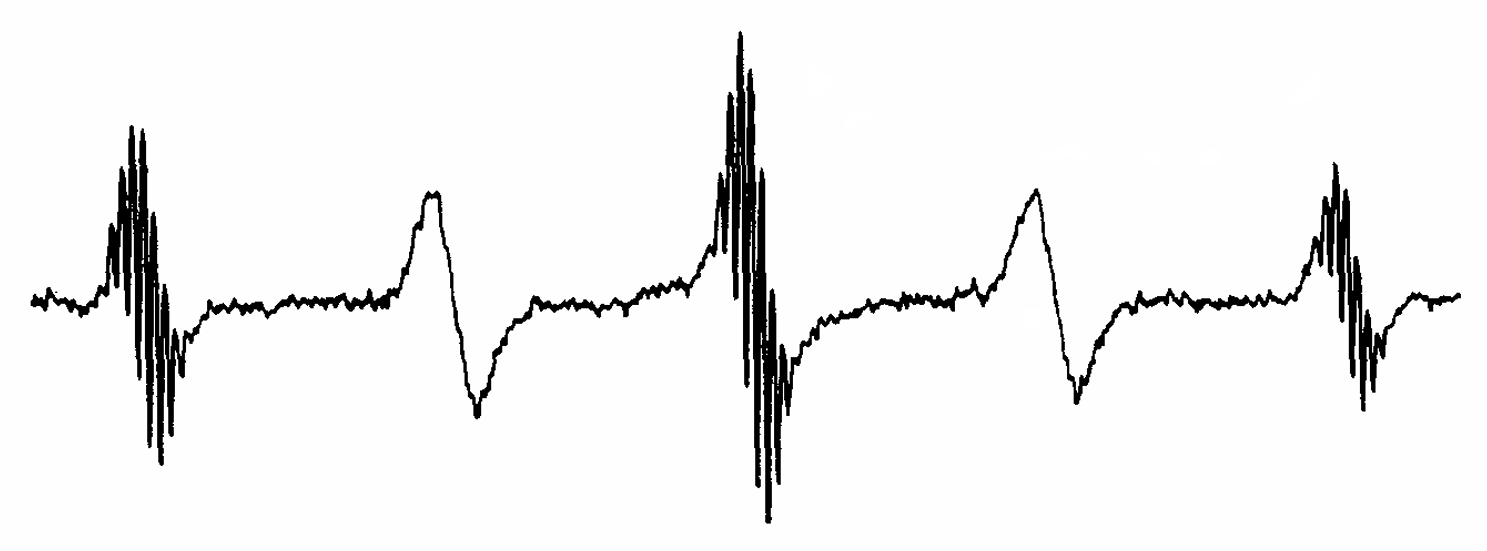
|
|
ABSTRACT: An anomalous alternation in the widths of the hyperfine components has been found in the ESR spectrum of the 1,4-dinitrotetramethylbenzene (dinitrodurene) anion radical. The radical was produced by electrolytic reduction of an N, N-dimethylformamide solution of dinitrodurene. The ESR spectrum, Fig. 1, shows a splitting into five major lines corresponding to hyperfine interactions with two equivalent nitrogen nuclei of aN = 6.99 G. The relative amplitudes of these lines clearly deviate from the predicted relative statistical weights of 1 : 2 : 3 : 2 : 1. The further splitting in the spectrum arises from interactions with the 12 equivalent methyl protons of aH = 0.25 G. The minor components of the 0 and ± 2 lines are comparable in resolution and width but are considerably better resolved and narrower than the ± 1 lines. These differences decrease with increasing temperature.
|
|
© 2022
|
.svg)

